YES1 (YES proto-oncogene 1, Src family tyrosine kinase)
- symbol:
- YES1
- locus group:
- protein-coding gene
- location:
- 18p11.32
- gene_family:
- SH2 domain containing
- alias symbol:
- Yes|c-yes|HsT441
- alias name:
- None
- entrez id:
- 7525
- ensembl gene id:
- ENSG00000176105
- ucsc gene id:
- uc002kkz.4
- refseq accession:
- NM_005433
- hgnc_id:
- HGNC:12841
- approved reserved:
- 1986-01-01
YES1基因属于SRC基因家族,这是一个编码非受体型酪氨酸激酶的家族,其成员在细胞信号转导中起关键作用,通过磷酸化靶蛋白调控细胞增殖、分化、迁移和存活。YES1编码的蛋白与SRC激酶结构相似,包含SH3(调控蛋白相互作用)、SH2(结合磷酸化酪氨酸)和激酶结构域(催化磷酸化反应),主要定位于细胞膜内侧和细胞黏附连接处,参与整合素介导的细胞黏附信号和生长因子受体(如EGFR)下游通路。该基因的激活依赖C端酪氨酸残基(Y537)的去磷酸化,而激酶活性受CSK等激酶的抑制性磷酸化调控。功能上,YES1在胚胎发育中调节组织形态发生,成年后在上皮细胞、神经元和免疫细胞中维持稳态,例如促进紧密连接形成、突触可塑性及B细胞活化。突变可能导致功能获得(如激酶区突变增强活性)或功能丧失(如截短突变),前者与肿瘤发生相关(如结肠癌、肺癌中YES1扩增或突变导致增殖和侵袭增强),后者可能引起发育缺陷。与疾病关联方面,YES1过表达常见于乳腺癌、头颈癌等恶性肿瘤,通过激活PI3K-AKT和STAT3通路促进存活和转移;而敲低YES1可抑制肿瘤生长但可能影响正常组织修复。基因家族共性上,SRC家族成员(如SRC、LYN、FYN)均通过调控细胞骨架动力学和生长信号参与癌症,且共享自抑制调节机制。表达异常时,YES1过表达会增强下游ERK和FAK信号,导致异常增殖;低表达则可能削弱细胞迁移能力。需要注意的是,中文文献中"YES1"偶被译为"耶斯1",但统一使用"YES1"更规范。
This gene is the cellular homolog of the Yamaguchi sarcoma virus oncogene. The encoded protein has tyrosine kinase activity and belongs to the src family of proteins. This gene lies in close proximity to thymidylate synthase gene on chromosome 18, and a corresponding pseudogene has been found on chromosome 22. [provided by RefSeq, Jul 2008]
此基因是山口肉瘤病毒原癌基因的细胞同系物。所编码的蛋白质具有酪氨酸激酶活性和属于src家族蛋白质。该基因位于靠近胸苷酸合成酶基因在染色体18和对应的假基因已在[通过的RefSeq,2008年7月提供]染色体22被发现
基因本体信息
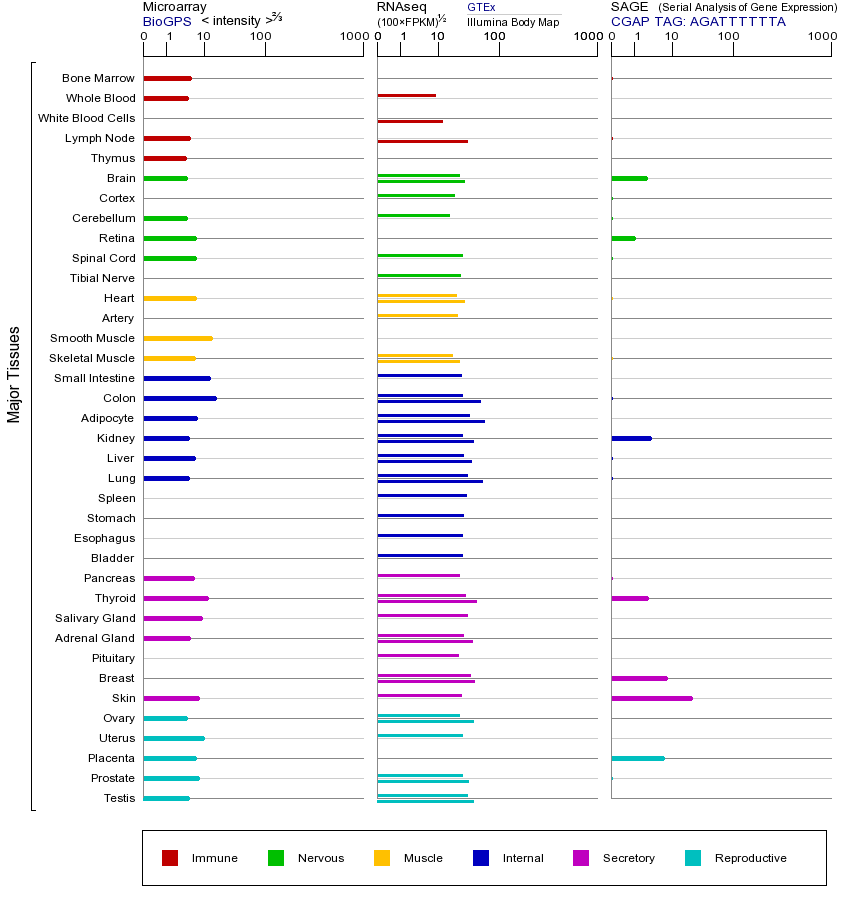
YES1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
YES1基因的本体(GO)信息:
| 名称 |
|---|
| 4520 Adherens junction [PATH:hsa04520] |
| 4530 Tight junction [PATH:hsa04530] |
| 名称 |
|---|
| Adaptive Immune System |
| Axon guidance |
| CD28 co-stimulation |
| Cell surface interactions at the vascular wall |
| Costimulation by the CD28 family |
| CTLA4 inhibitory signaling |
| Cytokine Signaling in Immune system |
| Developmental Biology |
| EPH-ephrin mediated repulsion of cells |
| EPH-Ephrin signaling |
| EPHA-mediated growth cone collapse |
| EPHB-mediated forward signaling |
| Fcgamma receptor (FCGR) dependent phagocytosis |
| FCGR activation |
| Hemostasis |
| Innate Immune System |
| Interleukin-3, 5 and GM-CSF signaling |
| PECAM1 interactions |
| Regulation of KIT signaling |
| Regulation of signaling by CBL |
| Signaling by ERBB2 |
| Signaling by Interleukins |
| Signaling by SCF-KIT |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| IGA Glomerulonephritis | 0.12 | 1 | 0 | CTD_human |
| Neoplasm Metastasis | 0.005720142 | 2 | 0 | BeFree_LHGDN |
| Glioblastoma | 0.00272435 | 1 | 0 | LHGDN |
| Mammary Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
| Stomach Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
| Colorectal Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
| Liver carcinoma | 0.00272435 | 1 | 0 | LHGDN |
| HIV Infections | 0.002367032 | 1 | 0 | GAD |
| Malignant neoplasm of stomach | 0.000542884 | 2 | 0 | BeFree |
| Stomach Carcinoma | 0.000542884 | 2 | 0 | BeFree |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。