TRPV4 (transient receptor potential cation channel subfamily V member 4)
- symbol:
- TRPV4
- locus group:
- protein-coding gene
- location:
- 12q24.11
- gene_family:
- Transient receptor potential cation channels|Ankyrin repeat domain containing
- alias symbol:
- OTRPC4|TRP12|VROAC|VRL-2|VR-OAC|CMT2C
- alias name:
- osmosensitive transient receptor p…
- entrez id:
- 59341
- ensembl gene id:
- ENSG00000111199
- ucsc gene id:
- uc001tpk.3
- refseq accession:
- NM_021625
- hgnc_id:
- HGNC:18083
- approved reserved:
- 2002-01-29
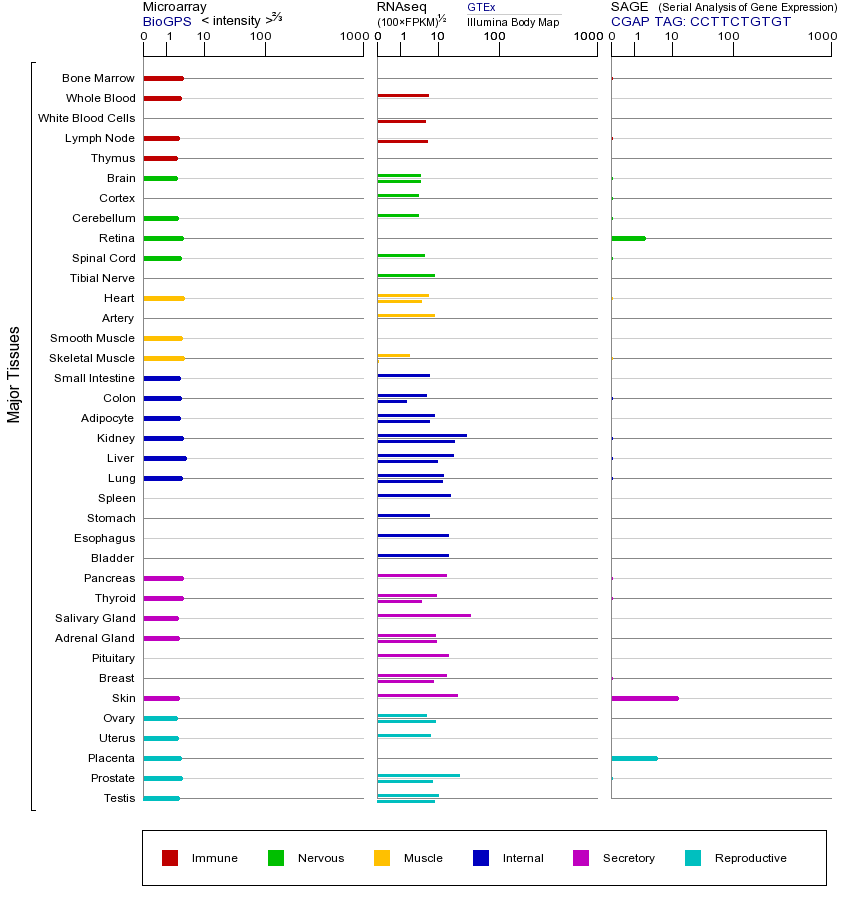
TRPV4(瞬时受体电位香草素4)是一种非选择性阳离子通道蛋白,属于TRP(瞬时受体电位)基因家族中的TRPV亚家族。TRPV4主要在感觉神经元、内皮细胞、肾脏、皮肤和软骨等组织中表达,参与多种生理功能,包括温度感知(27-34°C的温暖范围)、机械刺激响应(如渗透压和牵张力)以及钙离子平衡调节。TRPV4通过感知细胞外环境变化而被激活,其激活机制包括热、低渗透压、内源性脂质介质(如花生四烯酸代谢物)以及剪切力等物理化学刺激。TRPV4的功能异常与多种疾病相关,例如突变可导致遗传性神经病变(如Charcot-Marie-Tooth病2C型)、骨骼发育异常(如先天性脊柱骨骺发育不良)和疼痛综合征。TRPV4突变通常导致通道功能增强(增益性突变),引发钙离子内流异常,进而影响细胞稳态。过表达TRPV4可能加剧炎症反应、疼痛敏感性和组织损伤,例如在关节炎或肺水肿中观察到其表达上调;而降低表达或抑制TRPV4则可能减轻机械性痛觉过敏和某些水肿症状。TRPV家族(TRPV1-6)的共性在于均包含6个跨膜结构域,形成非选择性阳离子通道,主要通透钙离子,并参与感知温度、渗透压和化学配体等刺激。TRPV4与家族其他成员(如TRPV1感知高温和辣椒素)功能部分重叠但具有独特的激活阈值和组织分布。此外,TRPV4与细胞骨架蛋白(如微管和肌动蛋白)相互作用,影响细胞形态和迁移,在血管舒张和骨发育中起关键作用。研究还发现TRPV4与机械敏感离子通道Piezo存在功能交叉,共同调控机械转导过程。
This gene encodes a member of the OSM9-like transient receptor potential channel (OTRPC) subfamily in the transient receptor potential (TRP) superfamily of ion channels. The encoded protein is a Ca2+-permeable, nonselective cation channel that is thought to be involved in the regulation of systemic osmotic pressure. Mutations in this gene are the cause of spondylometaphyseal and metatropic dysplasia and hereditary motor and sensory neuropathy type IIC. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2010]
这个基因编码离子通道的瞬时受体电位(TRP)家族的OSM9样瞬时受体电位通道(OTRPC)亚科的成员。所编码的蛋白质是一个钙离子-permeable,非选择性阳离子通道,被认为是参与全身渗透压的调节。在这个基因的突变是spondylometaphyseal和metatropic发育不良和遗传性运动感觉神经病变类型IIC的原因。已发现该基因编码不同亚型的多个抄本变形。 [由RefSeq的,2010年4月提供]
基因本体信息
TRPV4基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
TRPV4基因的本体(GO)信息:
| 名称 |
|---|
| 4750 Inflammatory mediator regulation of TRP channels [PATH:hsa04750] |
| 名称 |
|---|
| Ion channel transport |
| Stimuli-sensing channels |
| Transmembrane transport of small molecules |
| TRP channels |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Metatropic dwarfism | 0.482442977 | 9 | 9 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Brachyolmia Type 3 | 0.481628651 | 6 | 2 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Spondylometaphyseal dysplasia, Kozlowski type | 0.481357209 | 6 | 5 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Parastremmatic dwarfism | 0.480814326 | 4 | 1 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Scapuloperoneal Form of Spinal Muscular Atrophy | 0.480542884 | 3 | 3 | BeFree_CLINVAR_CTD_human_ORPHANET_UNIPROT |
| HEREDITARY MOTOR AND SENSORY NEUROPATHY, TYPE IIC (disorder) | 0.360542884 | 6 | 0 | BeFree_CTD_human_ORPHANET_UNIPROT |
| SPONDYLOEPIPHYSEAL DYSPLASIA, MAROTEAUX TYPE | 0.360271442 | 1 | 4 | BeFree_CLINVAR_CTD_human_ORPHANET |
| SPINAL MUSCULAR ATROPHY, DISTAL, CONGENITAL NONPROGRESSIVE (disorder) | 0.36 | 3 | 5 | CLINVAR_ORPHANET_UNIPROT |
| Digital Arthropathy-Brachydactyly, Familial | 0.36 | 1 | 3 | CLINVAR_ORPHANET_UNIPROT |
| Charcot-Marie-Tooth Disease | 0.241085767 | 5 | 4 | BeFree_CLINVAR_CTD_human |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。