POLB (DNA polymerase beta)
- symbol:
- POLB
- locus group:
- protein-coding gene
- location:
- 8p11.21
- gene_family:
- Polymerases, DNA-directed
- alias symbol:
- None
- alias name:
- None
- entrez id:
- 5423
- ensembl gene id:
- ENSG00000070501
- ucsc gene id:
- uc003xoz.3
- refseq accession:
- NM_002690
- hgnc_id:
- HGNC:9174
- approved reserved:
- 2001-06-22
POLB基因编码DNA聚合酶β(DNA polymerase beta),是哺乳动物细胞中负责碱基切除修复(BER)的关键酶之一。它属于X家族DNA聚合酶,该家族成员普遍具有较短的合成能力(通常仅合成1-10个核苷酸)但具有较高的保真度,主要参与DNA修复而非复制。POLB通过其两个结构域发挥作用:8kDa的N端结构域具有脱氧核糖磷酸二酯酶(dRP)裂解活性,而31kDa的C端结构域负责DNA合成。在BER途径中,POLB负责填补因损伤碱基切除后产生的单核苷酸缺口,这对维持基因组稳定性至关重要。POLB突变会导致BER功能缺陷,与多种癌症(如结直肠癌、前列腺癌)和神经系统疾病相关。某些突变(如R137Q、K167M)会显著降低酶活性,导致DNA损伤积累和细胞凋亡增加。POLB过表达常见于肿瘤细胞,可能通过错误修复促进基因组不稳定性;而敲除POLB会导致胚胎致死性,证明其在发育中的必要性。该基因还与化疗耐药性相关,某些肿瘤通过上调POLB来抵抗烷化剂类药物。X家族还包括其他成员如POLλ和POLμ,它们都具有类似的"手掌-手指-拇指"结构特征,但POLB是唯一在所有哺乳动物细胞中持续表达的成员。研究表明POLB与XRCC1、PARP1等修复蛋白形成功能复合体,其表达水平受p53等肿瘤抑制因子调控。
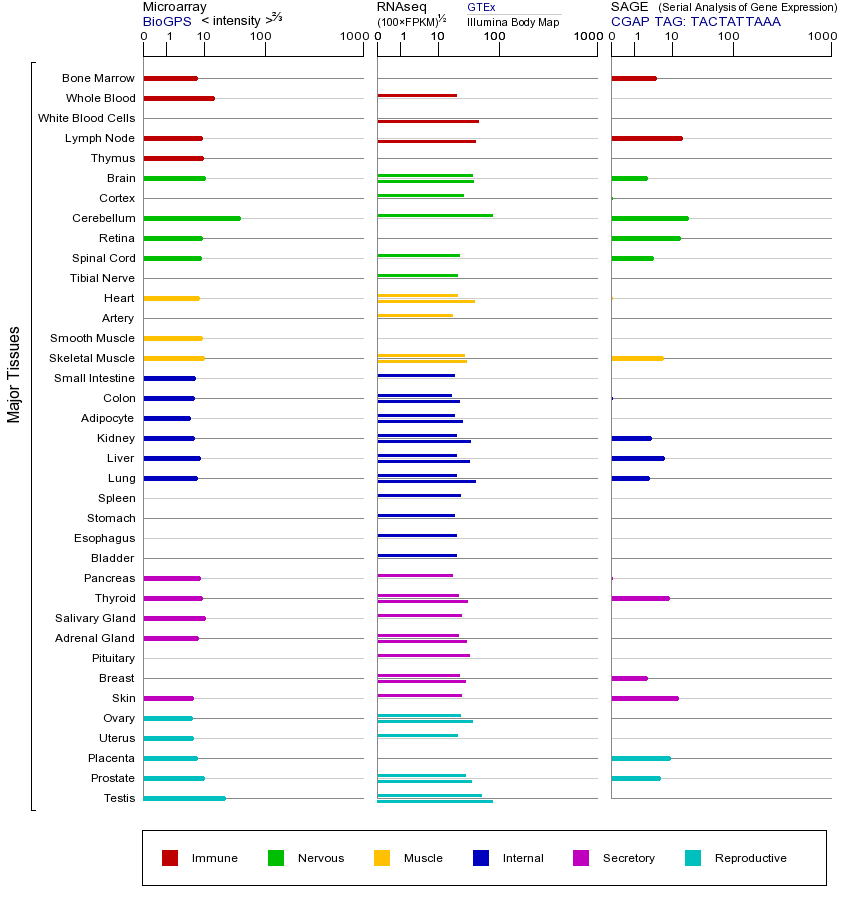
The protein encoded by this gene is a DNA polymerase involved in base excision and repair, also called gap-filling DNA synthesis. The encoded protein, acting as a monomer, is normally found in the cytoplasm, but it translocates to the nucleus upon DNA damage. Several transcript variants of this gene exist, but the full-length nature of only one has been described to date. [provided by RefSeq, Sep 2011]
由该基因编码的蛋白质是参与碱基切除和修理,也称为间隙填充DNA合成的DNA聚合酶。所编码的蛋白质,作为一个单体,通常是在细胞质中发现的,但它易位至在DNA损伤细胞核。此基因的多个转录物变体存在,但只有一个的全长性质已经描述到日期。 [由RefSeq的,2011年9月提供]
基因本体信息
POLB基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
POLB基因的本体(GO)信息:
| 名称 |
|---|
| 3410 Base excision repair [PATH:hsa03410] |
| 5203 Viral carcinogenesis [PATH:hsa05203] |
| 5166 HTLV-I infection [PATH:hsa05166] |
| 名称 |
|---|
| Abasic sugar-phosphate removal via the single-nucleotide replacement pathway |
| APEX1-Independent Resolution of AP Sites via the Single Nucleotide Replacement Pathway |
| Base Excision Repair |
| DNA Repair |
| PCNA-Dependent Long Patch Base Excision Repair |
| POLB-Dependent Long Patch Base Excision Repair |
| Resolution of Abasic Sites (AP sites) |
| Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| Resolution of AP sites via the single-nucleotide replacement pathway |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Chronic Lymphocytic Leukemia | 0.122367032 | 1 | 0 | CTD_human_GAD |
| Peripheral Neuropathy | 0.12 | 1 | 0 | CTD_human |
| Bladder Neoplasm | 0.12 | 1 | 0 | CTD_human |
| Lupus Erythematosus, Systemic | 0.08 | 0 | 0 | MGD |
| Reperfusion Injury | 0.08 | 1 | 0 | RGD |
| Malignant neoplasm of breast | 0.009001189 | 9 | 0 | BeFree_GAD |
| Malignant neoplasm of urinary bladder | 0.007915422 | 5 | 0 | BeFree_GAD |
| Malignant neoplasm of lung | 0.007372538 | 4 | 0 | BeFree_GAD |
| Colorectal Cancer | 0.005819831 | 6 | 0 | BeFree_GAD |
| Werner Syndrome | 0.003538676 | 4 | 0 | BeFree_LHGDN |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。