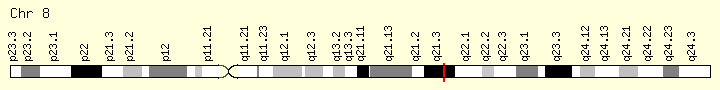NBN (nibrin)
- symbol:
- NBN
- locus group:
- protein-coding gene
- location:
- 8q21.3
- gene_family:
- alias symbol:
- ATV|AT-V2|AT-V1
- alias name:
- None
- entrez id:
- 4683
- ensembl gene id:
- ENSG00000104320
- ucsc gene id:
- uc003yej.2
- refseq accession:
- NM_001024688
- hgnc_id:
- HGNC:7652
- approved reserved:
- 1997-11-28
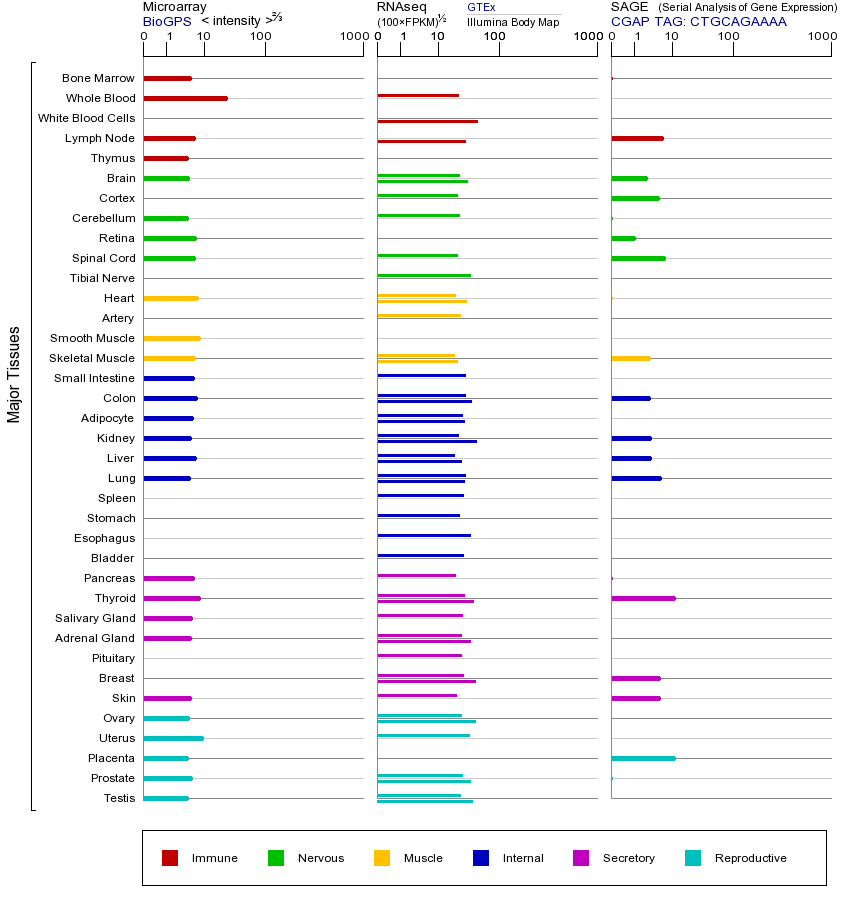
NBN基因(Nibrin)位于人类染色体8q21.3,编码一种称为nibrin的蛋白质,是MRE11-RAD50-NBN(MRN)复合体的核心成员之一。这个复合体在DNA双链断裂(DSB)修复和细胞周期检查点激活中起关键作用,特别是通过同源重组(HR)和非同源末端连接(NHEJ)途径修复DNA损伤。NBN蛋白直接参与识别和稳定断裂的DNA末端,并招募其他修复蛋白如ATM激酶,从而触发细胞对DNA损伤的反应。NBN基因突变会导致其功能丧失,影响MRN复合体的稳定性,进而损害DNA修复能力。最常见的NBN突变与奈梅亨断裂综合征(Nijmegen breakage syndrome, NBS)相关,这是一种常染色体隐性遗传病,患者表现为小头畸形、免疫缺陷、对辐射敏感以及癌症易感性(尤其是淋巴瘤)。此外,NBN突变还与乳腺癌、卵巢癌等癌症风险增加有关。如果NBN过表达,可能增强DNA修复能力,但异常高表达也可能干扰正常的细胞周期调控;而降低表达或功能缺失会导致基因组不稳定,增加突变积累和肿瘤发生风险。NBN属于MRE11/RAD50/NBN基因家族,这些基因的共性在于编码的蛋白质形成MRN复合体,共同参与DNA损伤应答、端粒维持和减数分裂重组。该家族成员在进化上高度保守,其功能缺陷均会导致基因组不稳定性及相关疾病。研究NBN不仅有助于理解DNA修复机制,还为癌症治疗(如靶向DNA修复途径的疗法)和遗传病诊断提供了重要线索。
Mutations in this gene are associated with Nijmegen breakage syndrome, an autosomal recessive chromosomal instability syndrome characterized by microcephaly, growth retardation, immunodeficiency, and cancer predisposition. The encoded protein is a member of the MRE11/RAD50 double-strand break repair complex which consists of 5 proteins. This gene product is thought to be involved in DNA double-strand break repair and DNA damage-induced checkpoint activation. [provided by RefSeq, Jul 2008]
在这个基因的突变与奈梅亨断裂综合征,一种常染色体隐性染色体不稳定综合征,表现为小头畸形,发育迟缓,免疫缺陷和癌症易感性有关。所编码的蛋白质是MRE11 / RAD50双链断裂修复它由5蛋白质的成员。该基因产物被认为是参与DNA双链断裂修复和DNA损伤诱导的检查点活化。 [由RefSeq的,2008年7月提供]
基因本体信息
NBN基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
NBN基因的本体(GO)信息:
| 名称 |
|---|
| 3440 Homologous recombination [PATH:hsa03440] |
| 名称 |
|---|
| Assembly of the RAD50-MRE11-NBS1 complex at DNA double-strand breaks |
| ATM mediated phosphorylation of repair proteins |
| ATM mediated response to DNA double-strand break |
| Cell Cycle |
| Cellular responses to stress |
| Cellular Senescence |
| DNA Damage/Telomere Stress Induced Senescence |
| DNA Repair |
| Double-Strand Break Repair |
| Homologous Recombination Repair |
| Homologous recombination repair of replication-independent double-strand breaks |
| Meiosis |
| Meiotic recombination |
| MRN complex relocalizes to nuclear foci |
| Recruitment of repair and signaling proteins to double-strand breaks |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Nijmegen Breakage Syndrome | 0.495682767 | 108 | 13 | BeFree_CLINVAR_CTD_human_GAD_LHGDN_MGD_ORPHANET |
| Malignant neoplasm of breast | 0.193953213 | 50 | 6 | BeFree_GAD_UNIPROT |
| Aplastic Anemia | 0.120542884 | 2 | 1 | BeFree_CLINVAR |
| Stomatitis | 0.120271442 | 1 | 0 | BeFree_CTD_human |
| Neoplastic Syndromes, Hereditary | 0.12 | 0 | 31 | CLINVAR |
| Prostatic Neoplasms | 0.12 | 1 | 0 | CTD_human |
| BREAST-OVARIAN CANCER, FAMILIAL, SUSCEPTIBILITY TO, 1 | 0.12 | 0 | 1 | CLINVAR |
| Hereditary Breast and Ovarian Cancer Syndrome | 0.12 | 0 | 0 | ORPHANET |
| Prostate cancer, familial | 0.12 | 0 | 0 | ORPHANET |
| Mammary Neoplasms | 0.024161834 | 9 | 0 | GAD_LHGDN |
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。