LIG1 (DNA ligase 1)
- symbol:
- LIG1
- locus group:
- protein-coding gene
- location:
- 19q13.33
- gene_family:
- alias symbol:
- LIGI|hLig1
- alias name:
- DNA ligase I
- entrez id:
- 3978
- ensembl gene id:
- ENSG00000105486
- ucsc gene id:
- uc002pia.2
- refseq accession:
- NM_000234
- hgnc_id:
- HGNC:6598
- approved reserved:
- 1991-05-09
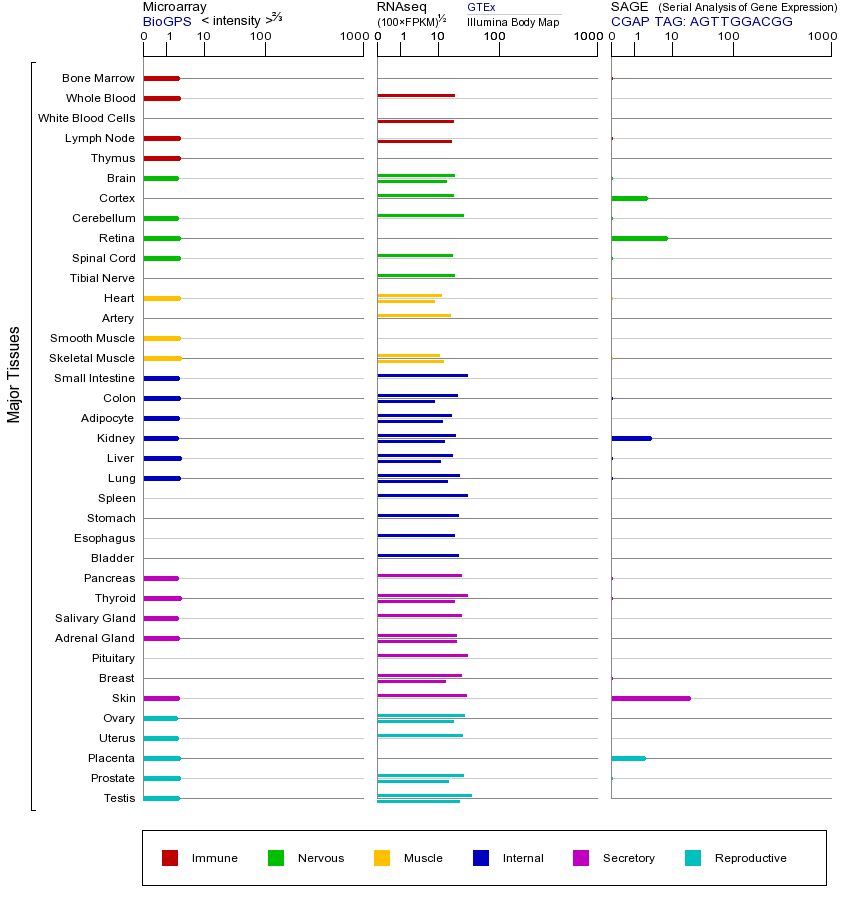
LIG1(DNA连接酶1)是一种关键的DNA修复酶,属于ATP依赖型DNA连接酶家族,主要功能是在DNA复制和修复过程中催化DNA链间的磷酸二酯键形成,确保DNA的完整性和稳定性。它在DNA单链断裂修复、碱基切除修复和核苷酸切除修复等过程中发挥核心作用,尤其在滞后链的Okazaki片段连接中至关重要。LIG1的表达产物是一种约102kDa的蛋白质,具有保守的催化结构域,能够识别并结合DNA缺口,利用ATP水解提供的能量完成连接反应。该基因的突变会导致多种严重后果,如LIG1综合征(表现为免疫缺陷、生长发育迟缓和光敏感性),部分突变还会增加癌症易感性(如结直肠癌和乳腺癌),因为DNA修复缺陷会导致基因组不稳定性。LIG1过表达可能干扰正常的DNA损伤应答,导致复制压力增加;而表达降低则会引起DNA损伤累积、细胞凋亡增加和复制叉停滞。作为DNA连接酶家族成员(还包括LIG3和LIG4),这些酶共享保守的催化机制,但具有不同的亚细胞定位和功能偏好——LIG1主要参与核DNA代谢,LIG3存在于线粒体,LIG4则专门用于非同源末端连接修复。该家族共性包括依赖ATP/Mg²⁺的催化活性、识别DNA缺口的核心结构域以及维持基因组稳定的基本功能。LIG1的表达受细胞周期调控,在S期达到高峰,与DNA复制需求同步。研究发现LIG1与多种癌症相关,其表达水平在某些肿瘤中异常,可能作为潜在的诊断标志物或治疗靶点。此外,LIG1与PCNA(增殖细胞核抗原)的相互作用对其在复制叉上的招募和功能发挥至关重要。
This gene encodes a member of the ATP-dependent DNA ligase protein family. The encoded protein functions in DNA replication, recombination, and the base excision repair process. Mutations in this gene that lead to DNA ligase I deficiency result in immunodeficiency and increased sensitivity to DNA-damaging agents. Disruption of this gene may also be associated with a variety of cancers. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2014]
该基因编码的ATP依赖性DNA连接酶蛋白家族的一个成员。在DNA复制,重组和碱基切除修复过程所编码的蛋白质的功能。突变该基因导致DNA连接酶I缺乏导致免疫缺陷和敏感性增加对DNA损伤剂。这个基因的破坏也可以与多种癌症相关联。选择性剪接结果在多个抄本变形。 [由RefSeq的,2014年1月提供]
基因本体信息
LIG1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
LIG1基因的本体(GO)信息:
| 名称 |
|---|
| 3030 DNA replication [PATH:hsa03030] |
| 3410 Base excision repair [PATH:hsa03410] |
| 3420 Nucleotide excision repair [PATH:hsa03420] |
| 3430 Mismatch repair [PATH:hsa03430] |
| 名称 |
|---|
| Base Excision Repair |
| Cell Cycle |
| Cell Cycle, Mitotic |
| Chromosome Maintenance |
| Disease |
| DNA Repair |
| DNA Replication |
| DNA strand elongation |
| Double-Strand Break Repair |
| Early Phase of HIV Life Cycle |
| Extension of Telomeres |
| Gap-filling DNA repair synthesis and ligation in GG-NER |
| Gap-filling DNA repair synthesis and ligation in TC-NER |
| Global Genomic NER (GG-NER) |
| HIV Infection |
| HIV Life Cycle |
| Homologous Recombination Repair |
| Homologous recombination repair of replication-independent double-strand breaks |
| Infectious disease |
| Lagging Strand Synthesis |
| Mismatch Repair |
| Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) |
| Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) |
| Nucleotide Excision Repair |
| PCNA-Dependent Long Patch Base Excision Repair |
| POLB-Dependent Long Patch Base Excision Repair |
| Processive synthesis on the C-strand of the telomere |
| Processive synthesis on the lagging strand |
| Resolution of Abasic Sites (AP sites) |
| Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| Resolution of D-loop structures |
| Resolution of D-loop structures through Holliday junction intermediates |
| S Phase |
| Synthesis of DNA |
| Telomere C-strand (Lagging Strand) Synthesis |
| Telomere Maintenance |
| Transcription-coupled NER (TC-NER) |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Bladder Neoplasm | 0.12 | 1 | 0 | CTD_human |
| Malignant neoplasm of lung | 0.007915422 | 6 | 0 | BeFree_GAD |
| Glioma | 0.007729856 | 2 | 2 | BeFree_GAD_LHGDN |
| Malignant neoplasm of breast | 0.007101096 | 3 | 0 | GAD |
| Malignant neoplasm of urinary bladder | 0.007101096 | 3 | 0 | GAD |
| Brain Neoplasms | 0.004734064 | 2 | 0 | GAD |
| Hematologic Neoplasms | 0.002367032 | 1 | 0 | GAD |
| Acoustic Neuroma | 0.002367032 | 1 | 0 | GAD |
| Lung Neoplasms | 0.002367032 | 1 | 0 | GAD |
| Graft-vs-Host Disease | 0.002367032 | 1 | 0 | GAD |
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。