INSR (insulin receptor)
- symbol:
- INSR
- locus group:
- protein-coding gene
- location:
- 19p13.2
- gene_family:
- Receptor Tyrosine Kinases|CD molecules|Fibronectin type III domain containing
- alias symbol:
- CD220
- alias name:
- None
- entrez id:
- 3643
- ensembl gene id:
- ENSG00000171105
- ucsc gene id:
- uc002mgd.2
- refseq accession:
- NM_000208
- hgnc_id:
- HGNC:6091
- approved reserved:
- 1986-01-01
INSR(胰岛素受体基因)编码胰岛素受体蛋白,是一种跨膜酪氨酸激酶受体,主要分布在肝脏、肌肉和脂肪组织等胰岛素敏感组织的细胞膜上。它的生物学功能是介导胰岛素信号传导,通过与胰岛素结合激活下游信号通路(如PI3K-AKT和MAPK通路),从而调控葡萄糖摄取、糖原合成、脂质代谢和细胞生长等关键生理过程。该基因表达产物为α和β亚基组成的异源四聚体,α亚基负责胰岛素结合,β亚基具有激酶活性。INSR属于受体酪氨酸激酶(RTK)基因家族,该家族成员均通过配体诱导的酪氨酸磷酸化传递细胞外信号。若INSR发生突变(如A1135T或R1152Q),可能导致受体功能丧失或信号传导障碍,引发胰岛素抵抗、Rabson-Mendenhall综合征或A型胰岛素抵抗等疾病。该基因过表达可能增强胰岛素敏感性,但异常高表达可能与某些癌症(如乳腺癌)的进展相关;而表达降低或功能缺陷则会导致高血糖、2型糖尿病或代谢综合征。INSR还与多囊卵巢综合征(PCOS)和阿尔茨海默病风险相关,因其信号通路影响神经元存活和tau蛋白磷酸化。该基因的剪接异构体INSR-A(缺失外显子11)主要在胎儿组织中表达,与细胞增殖相关;而INSR-B在成人组织中主导代谢调节。
After removal of the precursor signal peptide, the insulin receptor precursor is post-translationally cleaved into two chains (alpha and beta) that are covalently linked. Binding of insulin to the insulin receptor (INSR) stimulates glucose uptake. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008]
除去该前体的信号肽后,胰岛素受体前体的翻译后切割成两条链(α和β),该共价连接。结合胰岛素的胰岛素受体(INSR)刺激葡萄糖摄取。已发现该基因编码不同亚型的两个转录变异体。 [由RefSeq的,2008年7月提供]
基因本体信息
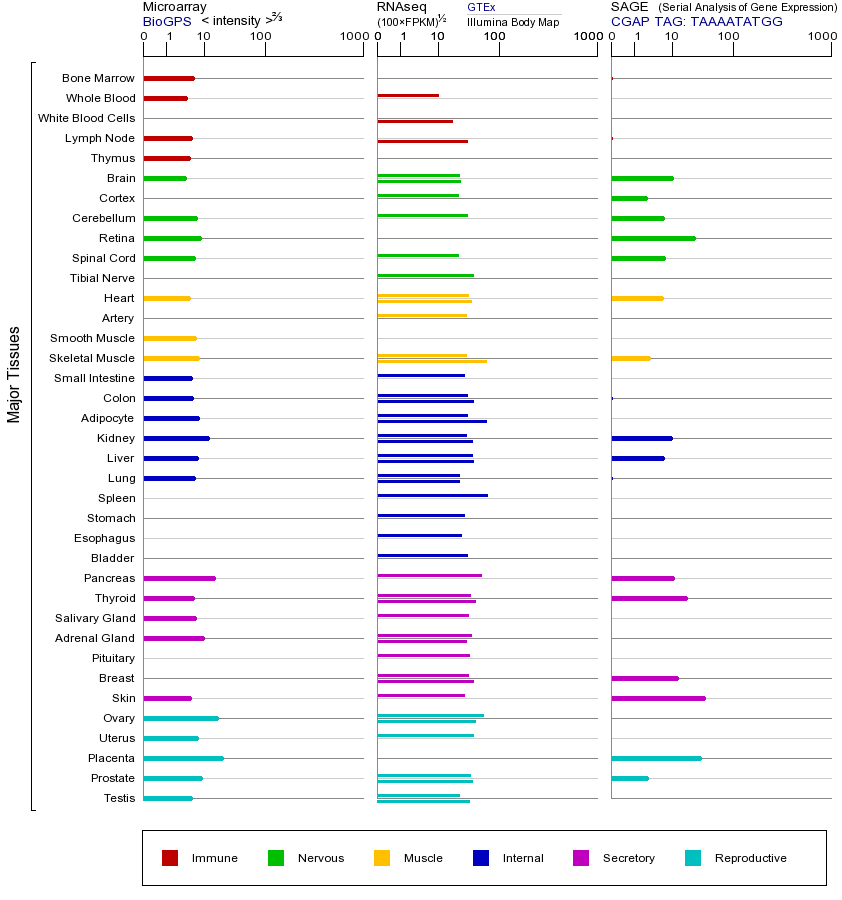
INSR基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
INSR基因的本体(GO)信息:
| 名称 |
|---|
| 4014 Ras signaling pathway [PATH:hsa04014] |
| 4015 Rap1 signaling pathway [PATH:hsa04015] |
| 4066 HIF-1 signaling pathway [PATH:hsa04066] |
| 4068 FoxO signaling pathway [PATH:hsa04068] |
| 4022 cGMP - PKG signaling pathway [PATH:hsa04022] |
| 4151 PI3K-Akt signaling pathway [PATH:hsa04151] |
| 4152 AMPK signaling pathway [PATH:hsa04152] |
| 4520 Adherens junction [PATH:hsa04520] |
| 4910 Insulin signaling pathway [PATH:hsa04910] |
| 4913 Ovarian Steroidogenesis [PATH:hsa04913] |
| 4960 Aldosterone-regulated sodium reabsorption [PATH:hsa04960] |
| 4930 Type II diabetes mellitus [PATH:hsa04930] |
| 4932 Non-alcoholic fatty liver disease (NAFLD) [PATH:hsa04932] |
| 名称 |
|---|
| Insulin receptor recycling |
| Insulin receptor signalling cascade |
| IRS activation |
| IRS-related events |
| SHC activation |
| SHC-related events |
| Signal attenuation |
| Signal Transduction |
| Signaling by Insulin receptor |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Donohue Syndrome | 0.493496149 | 46 | 17 | BeFree_CLINVAR_CTD_human_GAD_ORPHANET_UNIPROT |
| Hyperinsulinemic Hypoglycemia, Familial, 5 | 0.48 | 1 | 3 | CLINVAR_CTD_human_ORPHANET_UNIPROT |
| Diabetes Mellitus, Non-Insulin-Dependent | 0.401043039 | 114 | 11 | BeFree_CLINVAR_GAD_RGD_UNIPROT |
| Rabson-Mendenhall Syndrome | 0.364071628 | 19 | 4 | BeFree_CLINVAR_ORPHANET_UNIPROT |
| Insulin Resistance | 0.33183516 | 8 | 3 | CLINVAR_CTD_human_GAD_RGD |
| Diabetes Mellitus, Insulin-Resistant, with Acanthosis Nigricans | 0.24 | 18 | 5 | CTD_human_UNIPROT |
| Alzheimer's Disease | 0.200542884 | 4 | 0 | BeFree_CTD_human_RGD |
| Diabetic Neuropathies | 0.2 | 1 | 0 | CTD_human_RGD |
| Diabetes Mellitus, Experimental | 0.2 | 5 | 0 | CTD_human_RGD |
| Polycystic Ovary Syndrome | 0.159801061 | 34 | 2 | BeFree_GAD_GWASCAT_LHGDN |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。