HRAS (HRas proto-oncogene, GTPase)
- symbol:
- HRAS
- locus group:
- protein-coding gene
- location:
- 11p15.5
- gene_family:
- RAS type family GTPases
- alias symbol:
- None
- alias name:
- None
- entrez id:
- 3265
- ensembl gene id:
- ENSG00000174775
- ucsc gene id:
- uc010qvx.3
- refseq accession:
- NM_176795
- hgnc_id:
- HGNC:5173
- approved reserved:
- 2001-06-22
HRAS基因属于RAS基因家族,该家族还包括KRAS和NRAS,它们编码的蛋白质都是小GTP酶,在细胞信号传导中起关键作用,主要调控细胞生长、分化和存活。HRAS基因编码的H-Ras蛋白通过结合GTP(激活状态)和GDP(失活状态)来切换其功能状态,参与MAPK和PI3K-AKT等信号通路的调控。H-Ras蛋白的常见突变位点是第12、13或61位氨基酸,这些突变会导致GTP酶活性丧失,使蛋白持续处于激活状态,从而促进细胞异常增殖和肿瘤发生。HRAS的突变与多种癌症相关,如膀胱癌、甲状腺癌和头颈部鳞状细胞癌。HRAS基因的过表达会增强细胞增殖和存活信号,可能导致肿瘤发生和转移;而降低表达则可能抑制细胞生长或促进凋亡。HRAS基因的异常表达还会影响其他基因的功能,例如上调MYC和Cyclin D1等促增殖基因的表达。RAS基因家族的共性包括编码小GTP酶、参与细胞信号传导、在多种癌症中发生突变以及作为重要的治疗靶点。HRAS的功能研究为癌症治疗提供了潜在靶点,针对RAS信号通路的抑制剂正在开发中。
This gene belongs to the Ras oncogene family, whose members are related to the transforming genes of mammalian sarcoma retroviruses. The products encoded by these genes function in signal transduction pathways. These proteins can bind GTP and GDP, and they have intrinsic GTPase activity. This protein undergoes a continuous cycle of de- and re-palmitoylation, which regulates its rapid exchange between the plasma membrane and the Golgi apparatus. Mutations in this gene cause Costello syndrome, a disease characterized by increased growth at the prenatal stage, growth deficiency at the postnatal stage, predisposition to tumor formation, mental retardation, skin and musculoskeletal abnormalities, distinctive facial appearance and cardiovascular abnormalities. Defects in this gene are implicated in a variety of cancers, including bladder cancer, follicular thyroid cancer, and oral squamous cell carcinoma. Multiple transcript variants, which encode different isoforms, have been identified for this gene. [provided by RefSeq, Jul 2008]
该基因属于ras癌基因家族,其成员都涉及到哺乳动物肉瘤逆转录病毒的基因转化。由这些基因编码的产品中的信号转导途径发挥作用。这些蛋白质能结合GTP和GDP,他们有内在的GTP酶活性。这种蛋白质经受德和重新棕榈,调控其质膜和高尔基体之间的快速交换的一个连续的循环。突变该基因引起科斯特洛综合征的疾病特征在于通过在产前阶段增加的生长,生长缺陷在产后阶段,易感性肿瘤的形成,智力低下,皮肤和肌肉骨骼异常,独特的面部外观和心血管异常。在这个基因的缺陷涉及各种癌症,包括膀胱癌,甲状腺滤泡状癌和口腔鳞状细胞癌的。多个转录变异体,其编码不同同种型,已经确定了该基因。 [由RefSeq的,2008年7月提供]
基因本体信息
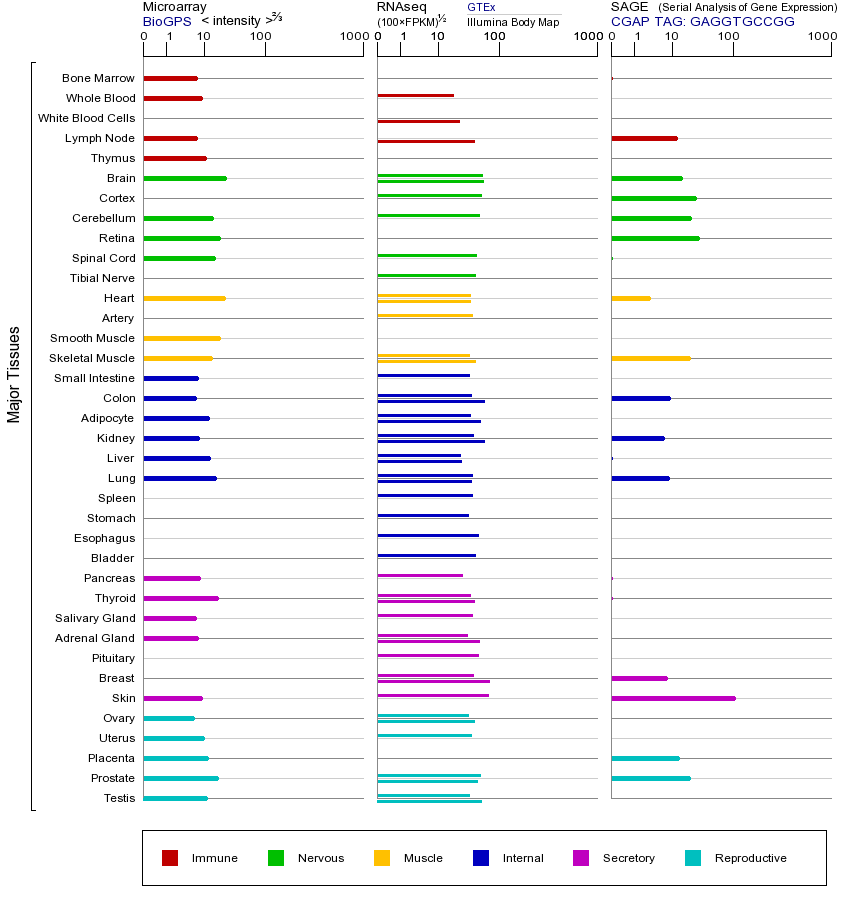
HRAS基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
HRAS基因的本体(GO)信息:
| 名称 |
|---|
| 4014 Ras signaling pathway [PATH:hsa04014] |
| 4015 Rap1 signaling pathway [PATH:hsa04015] |
| 4010 MAPK signaling pathway [PATH:hsa04010] |
| 4012 ErbB signaling pathway [PATH:hsa04012] |
| 4370 VEGF signaling pathway [PATH:hsa04370] |
| 4068 FoxO signaling pathway [PATH:hsa04068] |
| 4071 Sphingolipid signaling pathway [PATH:hsa04071] |
| 4151 PI3K-Akt signaling pathway [PATH:hsa04151] |
| 4144 Endocytosis [PATH:hsa04144] |
| 4810 Regulation of actin cytoskeleton [PATH:hsa04810] |
| 4510 Focal adhesion [PATH:hsa04510] |
| 4530 Tight junction [PATH:hsa04530] |
| 4540 Gap junction [PATH:hsa04540] |
| 4550 Signaling pathways regulating pluripotency of stem cells [PATH:hsa04550] |
| 4650 Natural killer cell mediated cytotoxicity [PATH:hsa04650] |
| 4660 T cell receptor signaling pathway [PATH:hsa04660] |
| 4662 B cell receptor signaling pathway [PATH:hsa04662] |
| 4664 Fc epsilon RI signaling pathway [PATH:hsa04664] |
| 4062 Chemokine signaling pathway [PATH:hsa04062] |
| 4910 Insulin signaling pathway [PATH:hsa04910] |
| 4912 GnRH signaling pathway [PATH:hsa04912] |
| 4915 Estrogen signaling pathway [PATH:hsa04915] |
| 4917 Prolactin signaling pathway [PATH:hsa04917] |
| 4921 Oxytocin signaling pathway [PATH:hsa04921] |
| 4919 Thyroid hormone signaling pathway [PATH:hsa04919] |
| 4916 Melanogenesis [PATH:hsa04916] |
| 4725 Cholinergic synapse [PATH:hsa04725] |
| 4726 Serotonergic synapse [PATH:hsa04726] |
| 4720 Long-term potentiation [PATH:hsa04720] |
| 4730 Long-term depression [PATH:hsa04730] |
| 4722 Neurotrophin signaling pathway [PATH:hsa04722] |
| 4360 Axon guidance [PATH:hsa04360] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5230 Central carbon metabolism in cancer [PATH:hsa05230] |
| 5231 Choline metabolism in cancer [PATH:hsa05231] |
| 5206 MicroRNAs in cancer [PATH:hsa05206] |
| 5205 Proteoglycans in cancer [PATH:hsa05205] |
| 5203 Viral carcinogenesis [PATH:hsa05203] |
| 5214 Glioma [PATH:hsa05214] |
| 5216 Thyroid cancer [PATH:hsa05216] |
| 5221 Acute myeloid leukemia [PATH:hsa05221] |
| 5220 Chronic myeloid leukemia [PATH:hsa05220] |
| 5218 Melanoma [PATH:hsa05218] |
| 5211 Renal cell carcinoma [PATH:hsa05211] |
| 5219 Bladder cancer [PATH:hsa05219] |
| 5215 Prostate cancer [PATH:hsa05215] |
| 5213 Endometrial cancer [PATH:hsa05213] |
| 5223 Non-small cell lung cancer [PATH:hsa05223] |
| 5034 Alcoholism [PATH:hsa05034] |
| 5166 HTLV-I infection [PATH:hsa05166] |
| 5161 Hepatitis B [PATH:hsa05161] |
| 5160 Hepatitis C [PATH:hsa05160] |
| 名称 |
|---|
| Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| Activation of RAS in B cells |
| Adaptive Immune System |
| ARMS-mediated activation |
| Axon guidance |
| C-type lectin receptors (CLRs) |
| CD209 (DC-SIGN) signaling |
| Cell surface interactions at the vascular wall |
| Constitutive Signaling by EGFRvIII |
| Constitutive Signaling by Ligand-Responsive EGFR Cancer Variants |
| CREB phosphorylation through the activation of Ras |
| Cytokine Signaling in Immune system |
| DAP12 interactions |
| DAP12 signaling |
| Developmental Biology |
| Disease |
| Diseases of signal transduction |
| Downstream signal transduction |
| Downstream signaling events of B Cell Receptor (BCR) |
| Downstream signaling of activated FGFR1 |
| Downstream signaling of activated FGFR2 |
| Downstream signaling of activated FGFR3 |
| Downstream signaling of activated FGFR4 |
| EGFR Transactivation by Gastrin |
| EPH-Ephrin signaling |
| EPHB-mediated forward signaling |
| Fc epsilon receptor (FCERI) signaling |
| FCERI mediated MAPK activation |
| FRS-mediated FGFR1 signaling |
| FRS-mediated FGFR2 signaling |
| FRS-mediated FGFR3 signaling |
| Frs2-mediated activation |
| FRS2-mediated FGFR4 signaling |
| Gastrin-CREB signalling pathway via PKC and MAPK |
| GRB2 events in EGFR signaling |
| GRB2 events in ERBB2 signaling |
| Hemostasis |
| IGF1R signaling cascade |
| Immune System |
| Innate Immune System |
| Insulin receptor signalling cascade |
| Interleukin receptor SHC signaling |
| Interleukin-2 signaling |
| Interleukin-3, 5 and GM-CSF signaling |
| IRS-mediated signalling |
| IRS-related events |
| IRS-related events triggered by IGF1R |
| MEK activation |
| NCAM signaling for neurite out-growth |
| Neuronal System |
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell |
| NGF signalling via TRKA from the plasma membrane |
| p38MAPK events |
| Post NMDA receptor activation events |
| Prolonged ERK activation events |
| RAF activation |
| RAF phosphorylates MEK |
| RAF/MAP kinase cascade |
| Ras activation uopn Ca2+ infux through NMDA receptor |
| SHC-mediated cascade:FGFR1 |
| SHC-mediated cascade:FGFR2 |
| SHC-mediated cascade:FGFR3 |
| SHC-mediated cascade:FGFR4 |
| SHC-mediated signalling |
| SHC-related events |
| SHC-related events triggered by IGF1R |
| SHC1 events in EGFR signaling |
| SHC1 events in ERBB2 signaling |
| SHC1 events in ERBB4 signaling |
| Signal Transduction |
| Signaling by EGFR |
| Signaling by EGFR in Cancer |
| Signaling by EGFRvIII in Cancer |
| Signaling by ERBB2 |
| Signaling by ERBB4 |
| Signaling by FGFR |
| Signaling by FGFR in disease |
| Signaling by FGFR1 |
| Signaling by FGFR1 in disease |
| Signaling by FGFR1 mutants |
| Signaling by FGFR2 |
| Signaling by FGFR2 in disease |
| Signaling by FGFR2 mutants |
| Signaling by FGFR3 |
| Signaling by FGFR3 in disease |
| Signaling by FGFR3 mutants |
| Signaling by FGFR4 |
| Signaling by FGFR4 in disease |
| Signaling by FGFR4 mutants |
| Signaling by GPCR |
| Signaling by Insulin receptor |
| Signaling by Interleukins |
| Signaling by Leptin |
| Signaling by Ligand-Responsive EGFR Variants in Cancer |
| Signaling by PDGF |
| Signaling by SCF-KIT |
| Signaling by the B Cell Receptor (BCR) |
| Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) |
| Signaling by VEGF |
| Signalling by NGF |
| Signalling to ERKs |
| Signalling to p38 via RIT and RIN |
| Signalling to RAS |
| SOS-mediated signalling |
| Tie2 Signaling |
| Transmission across Chemical Synapses |
| VEGFA-VEGFR2 Pathway |
| VEGFR2 mediated cell proliferation |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Costello syndrome (disorder) | 0.454657861 | 60 | 5 | BeFree_CTD_human_MGD_ORPHANET_UNIPROT |
| Nevus sebaceous | 0.36 | 1 | 0 | CTD_human_ORPHANET_UNIPROT |
| Mouth Neoplasms | 0.202367032 | 3 | 0 | CTD_human_GAD_RGD |
| Mammary Neoplasms, Experimental | 0.2 | 3 | 0 | CTD_human_RGD |
| Thyroid Neoplasm | 0.135731154 | 10 | 0 | BeFree_CTD_human_GAD_LHGDN |
| Squamous cell carcinoma | 0.130867606 | 31 | 0 | BeFree_CTD_human_LHGDN |
| Mammary Neoplasms | 0.127620235 | 14 | 0 | BeFree_CTD_human_LHGDN |
| Papilloma | 0.125710211 | 12 | 0 | BeFree_CTD_human_LHGDN |
| Bladder Neoplasm | 0.124538567 | 12 | 1 | BeFree_CTD_human_GAD |
| Carcinoma, Transitional Cell | 0.123452799 | 6 | 0 | BeFree_CTD_human_GAD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。