FYN (FYN proto-oncogene, Src family tyrosine kinase)
- symbol:
- FYN
- locus group:
- protein-coding gene
- location:
- 6q21
- gene_family:
- SH2 domain containing
- alias symbol:
- SYN|SLK|MGC45350
- alias name:
- None
- entrez id:
- 2534
- ensembl gene id:
- ENSG00000010810
- ucsc gene id:
- uc003pvk.3
- refseq accession:
- NM_001370529
- hgnc_id:
- HGNC:4037
- approved reserved:
- 1988-08-19
FYN是一种属于SRC家族的非受体型酪氨酸激酶基因,在细胞信号传导中发挥重要作用。该基因编码的Fyn蛋白广泛分布于多种组织中,尤其在神经系统和免疫系统中表达丰富。Fyn蛋白通过其SH2和SH3结构域与其他蛋白质相互作用,参与调控T细胞受体信号通路、神经元发育、突触可塑性以及细胞黏附等关键生物学过程。在免疫系统中,Fyn与Lck协同作用,参与T细胞激活的早期信号事件。在神经系统中,Fyn通过调节NMDA受体和髓鞘形成影响学习记忆功能。FYN基因突变可能导致多种疾病,包括神经系统疾病如阿尔茨海默病(通过影响tau蛋白磷酸化)和免疫系统异常。FYN过表达与某些癌症如乳腺癌和前列腺癌的进展相关,可能促进肿瘤细胞迁移和侵袭;而表达降低则可能损害T细胞功能导致免疫缺陷。FYN属于SRC基因家族,该家族成员均含有SH2、SH3结构域和激酶结构域,通过酪氨酸磷酸化参与细胞信号转导,在细胞生长、分化、存活和运动等过程中起调控作用。SRC家族激酶通常通过与细胞膜受体结合而被激活,其活性异常与多种癌症发生密切相关。
This gene is a member of the protein-tyrosine kinase oncogene family. It encodes a membrane-associated tyrosine kinase that has been implicated in the control of cell growth. The protein associates with the p85 subunit of phosphatidylinositol 3-kinase and interacts with the fyn-binding protein. Alternatively spliced transcript variants encoding distinct isoforms exist. [provided by RefSeq, Jul 2008]
该基??因是蛋白质 - 酪氨酸激酶致癌基因家族的一个成员。它编码已在细胞生长的控制被牵连的膜相关的酪氨酸激酶。与磷脂酰肌醇3激酶的P85亚基蛋白联营公司及与菲英岛结合蛋白相互作用。存在选择性剪接转录变异体的编码不同的亚型。 [由RefSeq的,2008年7月提供]
基因本体信息
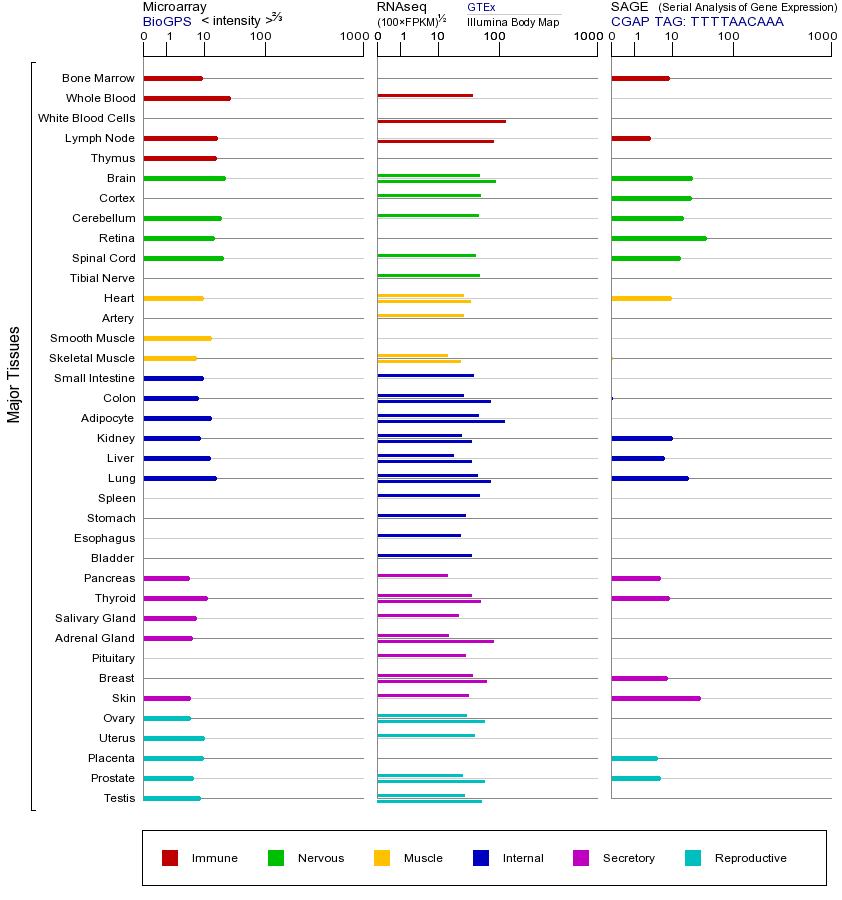
FYN基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
FYN基因的本体(GO)信息:
| 名称 |
|---|
| 4071 Sphingolipid signaling pathway [PATH:hsa04071] |
| 4510 Focal adhesion [PATH:hsa04510] |
| 4520 Adherens junction [PATH:hsa04520] |
| 4611 Platelet activation [PATH:hsa04611] |
| 4650 Natural killer cell mediated cytotoxicity [PATH:hsa04650] |
| 4660 T cell receptor signaling pathway [PATH:hsa04660] |
| 4664 Fc epsilon RI signaling pathway [PATH:hsa04664] |
| 4725 Cholinergic synapse [PATH:hsa04725] |
| 4360 Axon guidance [PATH:hsa04360] |
| 4380 Osteoclast differentiation [PATH:hsa04380] |
| 5020 Prion diseases [PATH:hsa05020] |
| 5416 Viral myocarditis [PATH:hsa05416] |
| 5130 Pathogenic Escherichia coli infection [PATH:hsa05130] |
| 5162 Measles [PATH:hsa05162] |
| 名称 |
|---|
| Adaptive Immune System |
| Antigen activates B Cell Receptor (BCR) leading to generation of second messengers |
| Axon guidance |
| C-type lectin receptors (CLRs) |
| CD209 (DC-SIGN) signaling |
| CD28 co-stimulation |
| CD28 dependent PI3K/Akt signaling |
| CD28 dependent Vav1 pathway |
| Cell surface interactions at the vascular wall |
| Cell-Cell communication |
| Constitutive Signaling by Aberrant PI3K in Cancer |
| Costimulation by the CD28 family |
| CRMPs in Sema3A signaling |
| CTLA4 inhibitory signaling |
| Cytokine Signaling in Immune system |
| DAP12 interactions |
| DAP12 signaling |
| DCC mediated attractive signaling |
| Dectin-2 family |
| Developmental Biology |
| Disease |
| Diseases of signal transduction |
| Downstream signal transduction |
| Downstream signaling events of B Cell Receptor (BCR) |
| Downstream signaling of activated FGFR1 |
| Downstream signaling of activated FGFR2 |
| Downstream signaling of activated FGFR3 |
| Downstream signaling of activated FGFR4 |
| EPH-ephrin mediated repulsion of cells |
| EPH-Ephrin signaling |
| EPHA-mediated growth cone collapse |
| EPHB-mediated forward signaling |
| Ephrin signaling |
| Fc epsilon receptor (FCERI) signaling |
| Fcgamma receptor (FCGR) dependent phagocytosis |
| FCGR activation |
| GAB1 signalosome |
| GPVI-mediated activation cascade |
| Hemostasis |
| HIV Infection |
| Host Interactions of HIV factors |
| Immune System |
| Infectious disease |
| Innate Immune System |
| Interleukin-3, 5 and GM-CSF signaling |
| NCAM signaling for neurite out-growth |
| Nef and signal transduction |
| Nephrin interactions |
| Netrin mediated repulsion signals |
| Netrin-1 signaling |
| NGF signalling via TRKA from the plasma membrane |
| PECAM1 interactions |
| PI-3K cascade:FGFR1 |
| PI-3K cascade:FGFR2 |
| PI-3K cascade:FGFR3 |
| PI-3K cascade:FGFR4 |
| PI3K events in ERBB2 signaling |
| PI3K events in ERBB4 signaling |
| PI3K/AKT activation |
| PI3K/AKT Signaling in Cancer |
| PIP3 activates AKT signaling |
| Platelet activation, signaling and aggregation |
| Platelet Adhesion to exposed collagen |
| Regulation of KIT signaling |
| Regulation of signaling by CBL |
| Role of LAT2/NTAL/LAB on calcium mobilization |
| Sema3A PAK dependent Axon repulsion |
| SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| Semaphorin interactions |
| Signal Transduction |
| Signaling by EGFR |
| Signaling by ERBB2 |
| Signaling by ERBB4 |
| Signaling by FGFR |
| Signaling by FGFR1 |
| Signaling by FGFR2 |
| Signaling by FGFR3 |
| Signaling by FGFR4 |
| Signaling by Interleukins |
| Signaling by PDGF |
| Signaling by SCF-KIT |
| Signaling by the B Cell Receptor (BCR) |
| Signaling by VEGF |
| Signalling by NGF |
| The role of Nef in HIV-1 replication and disease pathogenesis |
| VEGFA-VEGFR2 Pathway |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Peripheral T-Cell Lymphoma | 0.120271442 | 1 | 0 | BeFree_CTD_human |
| Stomach Neoplasms | 0.12 | 1 | 0 | CTD_human |
| Catalepsy | 0.12 | 1 | 0 | CTD_human |
| Adult T-Cell Lymphoma/Leukemia | 0.12 | 1 | 0 | CTD_human |
| Sepsis | 0.08 | 1 | 0 | RGD |
| Epilepsy, Temporal Lobe | 0.08 | 1 | 0 | RGD |
| Alzheimer's Disease | 0.010454206 | 5 | 0 | BeFree_GAD_LHGDN |
| Asthma | 0.005634266 | 2 | 5 | BeFree_GAD_LHGDN |
| Schizophrenia | 0.00554839 | 5 | 0 | BeFree_GAD |
| Bipolar Disorder | 0.005276948 | 2 | 0 | BeFree_GAD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。