EPAS1 (endothelial PAS domain protein 1)
- symbol:
- EPAS1
- locus group:
- protein-coding gene
- location:
- 2p21
- gene_family:
- Basic helix-loop-helix proteins
- alias symbol:
- MOP2|PASD2|HIF2A|HLF|bHLHe73
- alias name:
- HIF-1 alpha-like factor
- entrez id:
- 2034
- ensembl gene id:
- ENSG00000116016
- ucsc gene id:
- uc002ruv.3
- refseq accession:
- NM_001430
- hgnc_id:
- HGNC:3374
- approved reserved:
- 1998-05-29
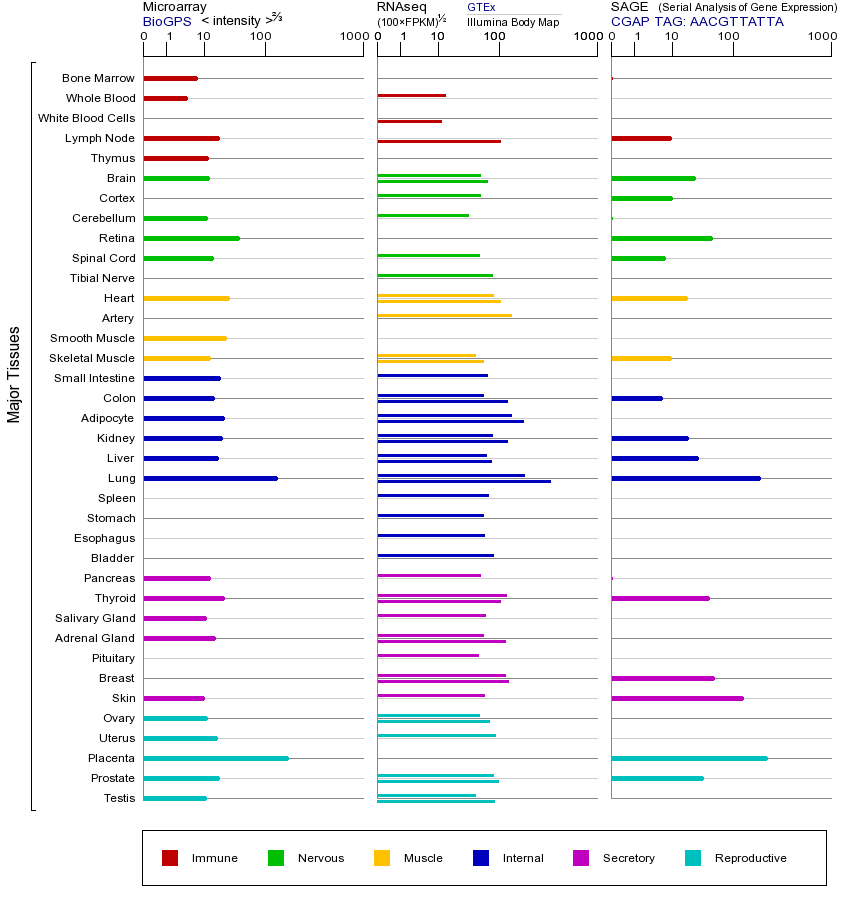
EPAS1(内皮PAS结构域蛋白1)基因,也称为HIF2A(缺氧诱导因子2α),属于缺氧诱导因子(HIF)基因家族。该家族包括HIF1A、HIF2A(EPAS1)和HIF3A,它们共同参与细胞对低氧环境的响应。HIF家族蛋白是转录因子,含有PAS(Per-ARNT-Sim)结构域和碱性螺旋-环-螺旋(bHLH)结构域,能与HIF1B(ARNT)结合形成异源二聚体,调控下游靶基因的表达,如促红细胞生成素(EPO)、血管内皮生长因子(VEGF)等,从而影响血管生成、红细胞生成和能量代谢。EPAS1主要在血管内皮细胞、肾脏和肝脏中表达,其功能与HIF1A类似但具有组织特异性,尤其在调节肾上腺髓质和肺血管发育中起关键作用。EPAS1的活性受氧浓度调控,在常氧条件下被脯氨酰羟化酶(PHD)羟基化,进而被泛素-蛋白酶体系统降解;而在缺氧条件下稳定表达并激活下游基因。EPAS1突变可导致多种疾病,如遗传性红细胞增多症(由于功能获得性突变使EPAS1持续激活,促进红细胞过度生成)或先天性肺动脉高压(功能缺失突变影响肺血管重塑)。此外,EPAS1的过表达与某些癌症(如嗜铬细胞瘤和副神经节瘤)的发生相关,因其促进血管生成和肿瘤微环境适应;而表达降低可能导致高原适应能力下降或贫血。EPAS1还与藏族人群的高原适应相关,其特定单核苷酸多态性(SNPs)能增强低氧耐受性。在基因家族中,HIF成员均通过氧敏感机制调控,但EPAS1相比HIF1A对慢性低氧的响应更持久,且在特定生理过程中(如胚胎发育)作用不可替代。
This gene encodes a transcription factor involved in the induction of genes regulated by oxygen, which is induced as oxygen levels fall. The encoded protein contains a basic-helix-loop-helix domain protein dimerization domain as well as a domain found in proteins in signal transduction pathways which respond to oxygen levels. Mutations in this gene are associated with erythrocytosis familial type 4. [provided by RefSeq, Nov 2009]
该基因编码参与由氧,氧水平下降而被诱导调控的基因的诱导的转录因子。所编码的蛋白质包含一个碱性螺旋 - 环 - 螺旋结构域蛋白的二聚化结构域以及在信号转导途径其中氧气含量做出反应蛋白质中发现的域。在这个基因的突变与[由RefSeq的,2009年11月提供]红细胞增多症家族型4相关
基因本体信息
EPAS1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
EPAS1基因的本体(GO)信息:
| 名称 |
|---|
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5211 Renal cell carcinoma [PATH:hsa05211] |
| 名称 |
|---|
| Cellular response to hypoxia |
| Cellular responses to stress |
| Developmental Biology |
| Developmental Biology |
| Oxygen-dependent asparagine hydroxylation of Hypoxia-inducible Factor Alpha |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha |
| Regulation of gene expression by Hypoxia-inducible Factor |
| Regulation of Hypoxia-inducible Factor (HIF) by oxygen |
| Transcriptional regulation of pluripotent stem cells |
| Transcriptional regulation of pluripotent stem cells |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Erythrocytosis, Familial, 4 | 0.36 | 4 | 2 | CLINVAR_CTD_human_UNIPROT |
| Renal Cell Carcinoma | 0.332701686 | 22 | 3 | BeFree_CTD_human_GAD_GWASCAT_LHGDN_RGD |
| Neoplastic Cell Transformation | 0.12 | 1 | 0 | CTD_human |
| Chromosome Aberrations | 0.12 | 1 | 0 | CTD_human |
| Pulmonary Hypertension | 0.083181358 | 5 | 0 | BeFree_GAD_RGD |
| Acute kidney injury | 0.08 | 1 | 0 | RGD |
| Infarction | 0.08 | 1 | 0 | RGD |
| Diabetes Mellitus, Non-Insulin-Dependent | 0.08 | 1 | 0 | RGD |
| Diabetic Nephropathy | 0.08 | 2 | 0 | RGD |
| Neuritis, Autoimmune, Experimental | 0.08 | 1 | 0 | RGD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。