DRD2 (dopamine receptor D2)
- symbol:
- DRD2
- locus group:
- protein-coding gene
- location:
- 11q23.2
- gene_family:
- Dopamine receptors
- alias symbol:
- None
- alias name:
- None
- entrez id:
- 1813
- ensembl gene id:
- ENSG00000149295
- ucsc gene id:
- uc001poa.5
- refseq accession:
- NM_000795
- hgnc_id:
- HGNC:3023
- approved reserved:
- 1989-02-23
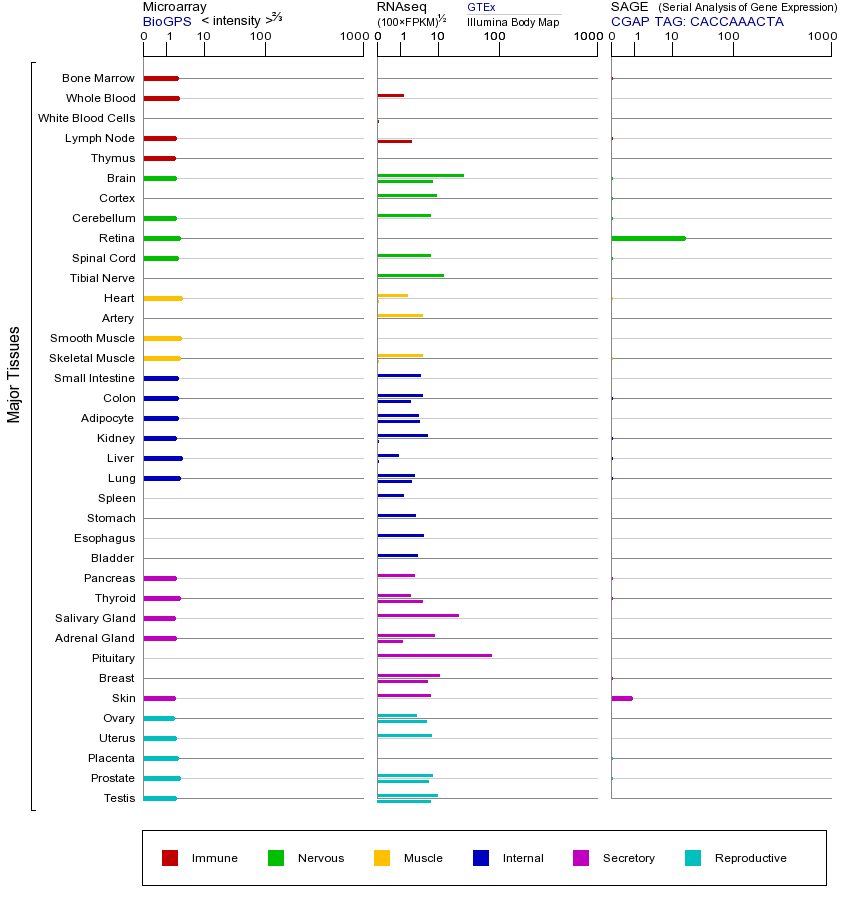
DRD2(多巴胺受体D2)基因编码多巴胺D2受体,属于G蛋白偶联受体家族中的多巴胺受体家族。该家族共有五种亚型(D1-D5),共同特点是介导多巴胺神经递质信号传导,参与运动控制、情绪调节和奖赏机制等生理过程。DRD2主要分布在大脑纹状体、伏隔核等区域,通过抑制腺苷酸环化酶活性来调节神经元兴奋性。其生物学功能包括调控运动协调性、认知功能和药物成瘾行为,在精神分裂症和帕金森病中起关键作用。DRD2基因突变可能导致受体功能异常,如Taq1A多态性与精神分裂症风险增加相关,某些突变还会降低抗精神病药物疗效。该基因表达异常与多种神经精神疾病密切相关:表达不足可能引发帕金森病运动症状,而过表达则与精神分裂症阳性症状(如幻觉妄想)相关。DRD2还参与调节其他神经递质系统,其表达水平变化会影响谷氨酸能、GABA能神经元的活动。在药物成瘾中,DRD2表达下调会导致奖赏系统敏感性改变。该基因也是抗精神病药物(如氟哌啶醇)的主要靶点,通过阻断D2受体发挥作用。值得注意的是,DRD2存在两种剪接变体(D2长型和短型),它们在信号传导效率和分布位置上存在差异,这种多样性增加了其在神经系统中的功能复杂性。
This gene encodes the D2 subtype of the dopamine receptor. This G-protein coupled receptor inhibits adenylyl cyclase activity. A missense mutation in this gene causes myoclonus dystonia; other mutations have been associated with schizophrenia. Alternative splicing of this gene results in two transcript variants encoding different isoforms. A third variant has been described, but it has not been determined whether this form is normal or due to aberrant splicing. [provided by RefSeq, Jul 2008]
该基因编码多巴胺受体的D2亚型。偶联受体该G蛋白抑制腺苷酸环化酶活性。在这个基因的错义突变导致肌阵挛性肌张力障碍;其他突变已与精神分裂症有关。两个转录本基因导致选择性剪接变异体编码不同亚型。第三种变体进行了说明,但它尚未确定这种形式是正常还是由于异常剪接。 [由RefSeq的,2008年7月提供]
基因本体信息
DRD2基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
DRD2基因的本体(GO)信息:
| 名称 |
|---|
| 4015 Rap1 signaling pathway [PATH:hsa04015] |
| 4024 cAMP signaling pathway [PATH:hsa04024] |
| 4080 Neuroactive ligand-receptor interaction [PATH:hsa04080] |
| 4540 Gap junction [PATH:hsa04540] |
| 4728 Dopaminergic synapse [PATH:hsa04728] |
| 5012 Parkinson's disease [PATH:hsa05012] |
| 5030 Cocaine addiction [PATH:hsa05030] |
| 5034 Alcoholism [PATH:hsa05034] |
| 名称 |
|---|
| Amine ligand-binding receptors |
| Class A/1 (Rhodopsin-like receptors) |
| Dopamine receptors |
| G alpha (i) signalling events |
| GPCR downstream signaling |
| GPCR ligand binding |
| Signal Transduction |
| Signaling by GPCR |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Schizophrenia | 0.291131658 | 162 | 12 | BeFree_CTD_human_GAD_LHGDN |
| Myoclonic dystonia | 0.241628651 | 6 | 0 | BeFree_CTD_human_ORPHANET |
| Parkinson Disease | 0.238958279 | 22 | 0 | BeFree_CTD_human_GAD_RGD |
| Cocaine-Related Disorders | 0.204734064 | 13 | 0 | CTD_human_GAD_RGD |
| Alcoholic Intoxication, Chronic | 0.203722004 | 163 | 7 | BeFree_GAD_LHGDN_RGD |
| Dyskinesia, Drug-Induced | 0.202367032 | 5 | 0 | CTD_human_GAD_RGD |
| Parkinsonian Disorders | 0.2 | 4 | 0 | CTD_human_RGD |
| Tachycardia | 0.2 | 3 | 0 | CTD_human_RGD |
| Attention deficit hyperactivity disorder | 0.155461872 | 34 | 5 | BeFree_CTD_human_GAD |
| Tobacco Use Disorder | 0.148404384 | 13 | 1 | CTD_human_GAD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。