CHEK1 (checkpoint kinase 1)
- symbol:
- CHEK1
- locus group:
- protein-coding gene
- location:
- 11q24.2
- gene_family:
- alias symbol:
- CHK1
- alias name:
- None
- entrez id:
- 1111
- ensembl gene id:
- ENSG00000149554
- ucsc gene id:
- uc001qcf.5
- refseq accession:
- NM_001274
- hgnc_id:
- HGNC:1925
- approved reserved:
- 1998-04-21
CHEK1(检查点激酶1)是一种关键的细胞周期调控基因,属于丝氨酸/苏氨酸激酶家族,与DNA损伤应答和细胞周期检查点密切相关。它主要在G2/M检查点发挥作用,当DNA受损时,CHEK1被上游激酶ATR磷酸化激活,进而磷酸化下游靶蛋白如CDC25C,阻止细胞进入有丝分裂,为DNA修复争取时间。CHEK1还参与S期检查点调控,确保DNA复制顺利完成。该基因的突变可能导致检查点功能缺陷,使细胞在DNA损伤情况下仍继续分裂,增加基因组不稳定性,与多种癌症如乳腺癌、卵巢癌和结直肠癌的发生发展相关。CHEK1过表达常见于某些肿瘤,可能导致对DNA损伤药物的耐药性,而降低表达则使细胞对辐射和化疗药物更敏感,因此它成为癌症治疗的潜在靶点。CHEK1属于检查点激酶家族,该家族还包括CHEK2,它们共同特点是参与DNA损伤应答,但CHEK1主要响应复制压力,而CHEK2更多参与双链断裂修复。CHEK1在胚胎发育中也很重要,完全敲除会导致小鼠胚胎致死,表明其对细胞存活不可或缺。此外,CHEK1与多种蛋白如BRCA1、p53等相互作用,形成复杂的调控网络。针对CHEK1的小分子抑制剂如MK-8776正在临床试验中,用于增强化疗效果。
The protein encoded by this gene belongs to the Ser/Thr protein kinase family. It is required for checkpoint mediated cell cycle arrest in response to DNA damage or the presence of unreplicated DNA. This protein acts to integrate signals from ATM and ATR, two cell cycle proteins involved in DNA damage responses, that also associate with chromatin in meiotic prophase I. Phosphorylation of CDC25A protein phosphatase by this protein is required for cells to delay cell cycle progression in response to double-strand DNA breaks. Several alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Oct 2011]
None
基因本体信息
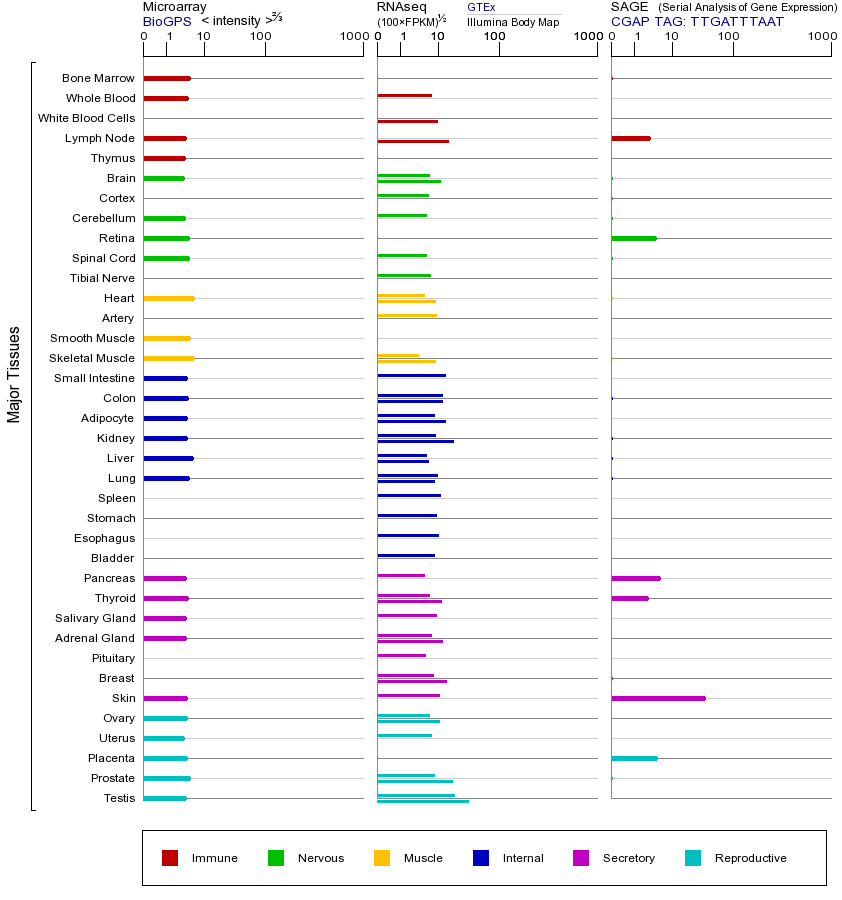
CHEK1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
CHEK1基因的本体(GO)信息:
| 名称 |
|---|
| 4110 Cell cycle [PATH:hsa04110] |
| 4115 p53 signaling pathway [PATH:hsa04115] |
| 5203 Viral carcinogenesis [PATH:hsa05203] |
| 5166 HTLV-I infection [PATH:hsa05166] |
| 名称 |
|---|
| Activation of ATR in response to replication stress |
| Cell Cycle |
| Cell Cycle Checkpoints |
| Chk1/Chk2(Cds1) mediated inactivation of Cyclin B:Cdk1 complex |
| G1/S DNA Damage Checkpoints |
| G2/M Checkpoints |
| G2/M DNA damage checkpoint |
| p53-Independent DNA Damage Response |
| p53-Independent G1/S DNA damage checkpoint |
| Signal Transduction |
| Signaling by SCF-KIT |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Mammary Neoplasms | 0.120542884 | 3 | 0 | BeFree_CTD_human |
| Polyploidy | 0.12 | 1 | 0 | CTD_human |
| Peripheral Neuropathy | 0.12 | 1 | 0 | CTD_human |
| IGA Glomerulonephritis | 0.12 | 1 | 0 | CTD_human |
| Malignant neoplasm of breast | 0.020173913 | 28 | 1 | BeFree_GAD |
| Fanconi Anemia | 0.009801702 | 9 | 0 | BeFree_LHGDN |
| Breast Carcinoma | 0.006243163 | 23 | 1 | BeFree |
| Malignant neoplasm of lung | 0.006091273 | 7 | 0 | BeFree_GAD |
| Colonic Neoplasms | 0.005091382 | 1 | 0 | GAD_LHGDN |
| DNA Damage | 0.004734064 | 2 | 0 | GAD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。