BID (BH3 interacting domain death agonist)
- symbol:
- BID
- locus group:
- protein-coding gene
- location:
- 22q11.21
- gene_family:
- Endogenous ligands
- alias symbol:
- None
- alias name:
- None
- entrez id:
- 637
- ensembl gene id:
- ENSG00000015475
- ucsc gene id:
- uc002znd.3
- refseq accession:
- NM_197966
- hgnc_id:
- HGNC:1050
- approved reserved:
- 1998-04-20
BID(BH3 interacting-domain death agonist)属于BH3-only蛋白家族,是Bcl-2蛋白家族的重要成员之一。该基因编码的蛋白是一种促凋亡蛋白,主要参与线粒体途径的细胞凋亡调控。BID的生物学功能是通过其BH3结构域(Bcl-2 homology domain 3)与其他Bcl-2家族蛋白相互作用,激活下游凋亡信号。当细胞受到凋亡刺激(如死亡受体激活)时,BID会被caspase-8剪切生成截短形式tBID(truncated BID),tBID转移到线粒体膜上,促使线粒体外膜通透性增加,导致细胞色素c释放,进而激活caspase级联反应引发细胞凋亡。BID的主要作用位点是线粒体,其功能受上游死亡受体信号(如Fas/CD95)调控。BID突变可能导致其无法被caspase-8有效剪切或丧失与Bcl-2家族蛋白相互作用的能力,从而抑制凋亡过程,这种异常与肿瘤发生发展相关。研究表明BID功能缺失可能促进肿瘤细胞存活,而其过度激活可能导致组织损伤。BID过表达会增强细胞对凋亡信号的敏感性,可能引起病理性细胞死亡;而低表达则可能导致凋亡抵抗,与癌症化疗耐药性有关。BID与多种疾病相关,包括肝癌、白血病等恶性肿瘤,以及肝衰竭等凋亡过度激活疾病。BH3-only蛋白家族的共性是通过BH3结构域与抗凋亡Bcl-2家族蛋白(如Bcl-2、Bcl-xL)结合,中和它们的抗凋亡作用或直接激活促凋亡蛋白(如BAX、BAK),在凋亡信号传递中起关键作用。这个家族的蛋白通常以非活性形式存在,需要被特定蛋白酶剪切或翻译后修饰才能发挥功能。BID的特殊性在于它是连接死亡受体通路和线粒体通路的关键分子。
This gene encodes a death agonist that heterodimerizes with either agonist BAX or antagonist BCL2. The encoded protein is a member of the BCL-2 family of cell death regulators. It is a mediator of mitochondrial damage induced by caspase-8 (CASP8); CASP8 cleaves this encoded protein, and the COOH-terminal part translocates to mitochondria where it triggers cytochrome c release. Multiple alternatively spliced transcript variants have been found, but the full-length nature of some variants has not been defined. [provided by RefSeq, Jul 2008]
None
基因本体信息
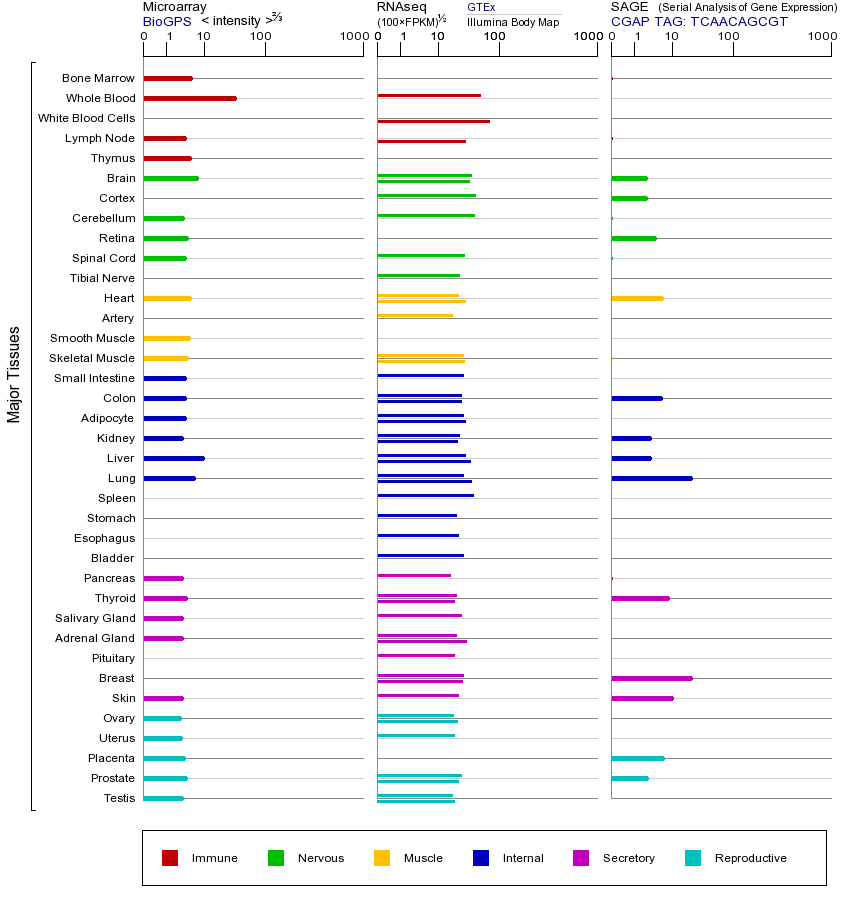
BID基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
BID基因的本体(GO)信息:
| 名称 |
|---|
| 4071 Sphingolipid signaling pathway [PATH:hsa04071] |
| 4210 Apoptosis [PATH:hsa04210] |
| 4115 p53 signaling pathway [PATH:hsa04115] |
| 4650 Natural killer cell mediated cytotoxicity [PATH:hsa04650] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5010 Alzheimer's disease [PATH:hsa05010] |
| 5014 Amyotrophic lateral sclerosis (ALS) [PATH:hsa05014] |
| 5416 Viral myocarditis [PATH:hsa05416] |
| 4932 Non-alcoholic fatty liver disease (NAFLD) [PATH:hsa04932] |
| 5152 Tuberculosis [PATH:hsa05152] |
| 名称 |
|---|
| Activation and oligomerization of BAK protein |
| Activation of BAD and translocation to mitochondria |
| Activation of BH3-only proteins |
| Activation, myristolyation of BID and translocation to mitochondria |
| Activation, translocation and oligomerization of BAX |
| Apoptosis |
| BH3-only proteins associate with and inactivate anti-apoptotic BCL-2 members |
| Intrinsic Pathway for Apoptosis |
| Programmed Cell Death |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Stomach Neoplasms | 0.122367032 | 2 | 0 | CTD_human_GAD |
| Liver carcinoma | 0.120271442 | 2 | 0 | BeFree_CTD_human |
| Myocardial Reperfusion Injury | 0.08 | 1 | 0 | RGD |
| Reperfusion Injury | 0.08 | 1 | 0 | RGD |
| Mammary Neoplasms | 0.008173051 | 3 | 0 | LHGDN |
| Adenocarcinoma | 0.007815732 | 3 | 0 | GAD_LHGDN |
| Malignant neoplasm of breast | 0.005819831 | 5 | 5 | BeFree_GAD |
| melanoma | 0.002995792 | 2 | 1 | BeFree_LHGDN |
| Colonic Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
| Lung Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。