BCAR1 (BCAR1 scaffold protein, Cas family member)
- symbol:
- BCAR1
- locus group:
- protein-coding gene
- location:
- 16q23.1
- gene_family:
- Cas scaffolding proteins
- alias symbol:
- P130Cas|Crkas|CAS|CASS1
- alias name:
- Crk-associated substrate|Cas scaff…
- entrez id:
- 9564
- ensembl gene id:
- ENSG00000050820
- ucsc gene id:
- uc002fdv.4
- refseq accession:
- NM_014567
- hgnc_id:
- HGNC:971
- approved reserved:
- 1999-04-29
BCAR1(Breast Cancer Anti-Estrogen Resistance 1)是一种多功能支架蛋白,主要参与细胞黏附、迁移和信号转导过程。它也被称为p130Cas(Crk-associated substrate),属于CAS(Crk-associated substrate)基因家族。该家族成员(如NEDD9、HEF1、EFS)的共同特点是含有多个蛋白相互作用结构域,如SH3结合域、底物结构域(SD)和富含脯氨酸的区域,这些结构域使其能够与多种信号分子结合,参与细胞骨架重组、增殖和存活等过程。BCAR1在整合素介导的信号通路中起关键作用,通过与Crk、FAK(黏着斑激酶)和SRC等蛋白相互作用,调控细胞外基质感知和细胞运动。突变或异常表达可能影响其功能,例如某些突变会导致BCAR1失去与Crk的结合能力,从而破坏下游信号传导,影响细胞迁移和侵袭。BCAR1的过表达与多种癌症相关,尤其是乳腺癌、肺癌和胶质母细胞瘤,因为它能增强肿瘤细胞的侵袭性和抗药性(如对抗雌激素治疗的抵抗)。此外,BCAR1高表达可能激活PI3K/AKT和MAPK通路,促进细胞存活和增殖。相反,降低BCAR1表达会抑制肿瘤转移,但可能影响正常细胞的黏附功能。在非病理状态下,BCAR1对胚胎发育、免疫细胞功能和血管生成也有重要作用。其表达水平受转录因子(如ETS1)和表观遗传调控影响。研究还发现BCAR1与炎症性疾病(如关节炎)相关,因其参与白细胞迁移。总体而言,BCAR1是细胞行为的关键调节因子,其功能失衡可能导致疾病,尤其是癌症进展。
BCAR1, or CAS, is an Src (MIM 190090) family kinase substrate involved in various cellular events, including migration, survival, transformation, and invasion (Sawada et al., 2006 [PubMed 17129785]).[supplied by OMIM, May 2009]
BCAR1,或CAS,是参与各种细胞的活动,包括迁移,存活,转化和侵袭中的Src(MIM 190090)家族激酶基板(泽田等,2006 [考研17129785])通过提供OMIM,2009年5月]
基因本体信息
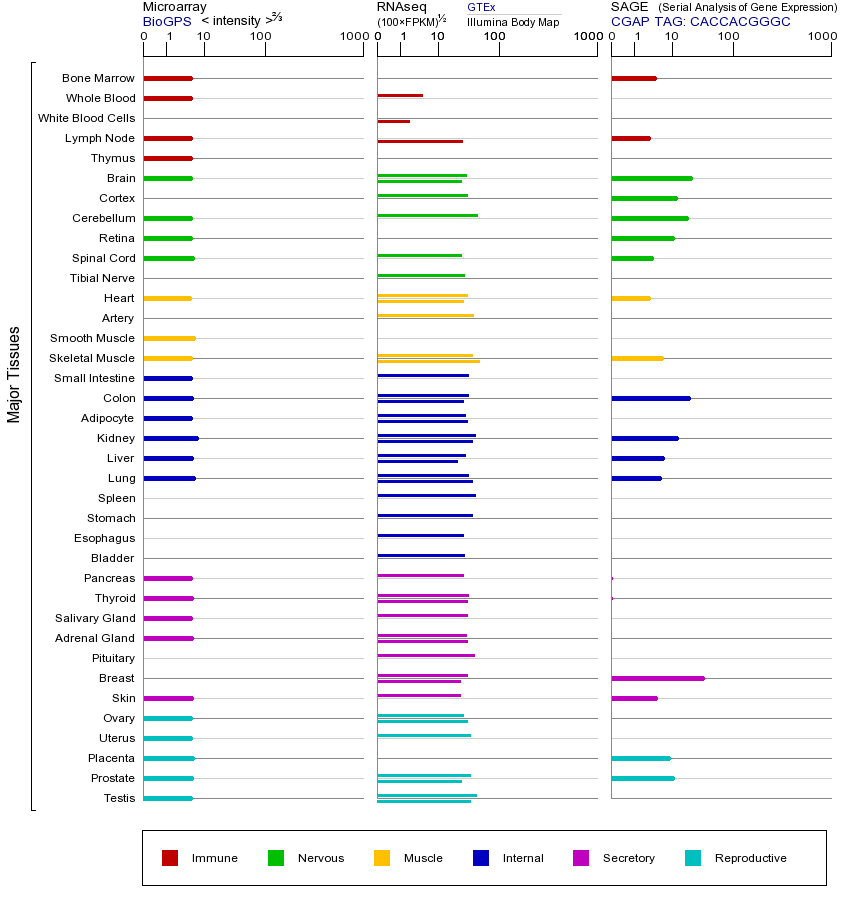
BCAR1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
BCAR1基因的本体(GO)信息:
| 名称 |
|---|
| 4015 Rap1 signaling pathway [PATH:hsa04015] |
| 4810 Regulation of actin cytoskeleton [PATH:hsa04810] |
| 4510 Focal adhesion [PATH:hsa04510] |
| 4670 Leukocyte transendothelial migration [PATH:hsa04670] |
| 4062 Chemokine signaling pathway [PATH:hsa04062] |
| 5100 Bacterial invasion of epithelial cells [PATH:hsa05100] |
| 名称 |
|---|
| Downstream signal transduction |
| Hemostasis |
| Integrin alphaIIb beta3 signaling |
| p130Cas linkage to MAPK signaling for integrins |
| Platelet activation, signaling and aggregation |
| Platelet Aggregation (Plug Formation) |
| Signal Transduction |
| Signaling by PDGF |
| Signaling by VEGF |
| VEGFA-VEGFR2 Pathway |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Pancreatic Neoplasm | 0.122367032 | 1 | 1 | CTD_human_GAD |
| Neoplasm Invasiveness | 0.12 | 1 | 0 | CTD_human |
| Malignant neoplasm of pancreas | 0.12 | 1 | 1 | GWASCAT |
| Mammary Neoplasms | 0.009801702 | 8 | 0 | BeFree_LHGDN |
| Malignant neoplasm of breast | 0.007328931 | 27 | 0 | BeFree |
| Breast Carcinoma | 0.006786047 | 25 | 0 | BeFree |
| melanoma | 0.006534468 | 4 | 0 | BeFree_LHGDN |
| Liver carcinoma | 0.003267234 | 3 | 0 | BeFree_LHGDN |
| Colonic Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
| Prostatic Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。