BAD (BCL2 associated agonist of cell death)
- symbol:
- BAD
- locus group:
- protein-coding gene
- location:
- 11q13.1
- gene_family:
- alias symbol:
- BCL2L8|BBC2
- alias name:
- None
- entrez id:
- 572
- ensembl gene id:
- ENSG00000002330
- ucsc gene id:
- uc001nzd.4
- refseq accession:
- NM_032989
- hgnc_id:
- HGNC:936
- approved reserved:
- 1997-10-16
BAD基因(BCL2-associated agonist of cell death)属于BCL-2蛋白家族,该家族成员主要调控细胞凋亡(程序性细胞死亡)过程。BAD基因编码的蛋白是一种促凋亡蛋白,通过与抗凋亡蛋白(如BCL-2和BCL-XL)结合并抑制其功能,从而促进细胞凋亡。BAD蛋白的活性受磷酸化调控,当被磷酸化时(如通过AKT或PKA激酶),它会与14-3-3蛋白结合并失去促凋亡活性;而去磷酸化则恢复其功能。BAD的主要作用位点是线粒体,它通过干扰BCL-2家族蛋白的平衡来触发线粒体外膜通透性增加,释放细胞色素C并激活凋亡信号通路。BAD基因突变可能导致其功能异常,例如失去促凋亡能力或持续激活,进而与癌症、神经退行性疾病等病理过程相关。若BAD过表达,可能增强细胞凋亡,导致组织损伤或发育异常;而降低表达则可能抑制凋亡,促进肿瘤细胞存活。BCL-2家族蛋白的共同特点是含有BH(BCL-2 homology)结构域,通过蛋白间相互作用调控凋亡。该家族包括促凋亡(如BAX、BAK)和抗凋亡(如BCL-2、BCL-XL)成员,共同维持细胞生死平衡。
The protein encoded by this gene is a member of the BCL-2 family. BCL-2 family members are known to be regulators of programmed cell death. This protein positively regulates cell apoptosis by forming heterodimers with BCL-xL and BCL-2, and reversing their death repressor activity. Proapoptotic activity of this protein is regulated through its phosphorylation. Protein kinases AKT and MAP kinase, as well as protein phosphatase calcineurin were found to be involved in the regulation of this protein. Alternative splicing of this gene results in two transcript variants which encode the same isoform. [provided by RefSeq, Jul 2008]
由该基因编码的蛋白质是BCL-2家族的一个成员。 BCL-2的FAMILY成员是已知的程序性细胞死亡的调节剂。该蛋白通过与BCL-xL和BCL-2的异二聚体,并且扭转他们??的死亡阻抑物活性正调节细胞凋亡。这种蛋白质的促凋亡活性是通过其磷酸化调节。蛋白激酶AKT和MAP激酶,以及蛋白磷酸酶钙调磷酸酶被发现参与该蛋白质的调节。该基因导致两个转录变体,其编码相同的同种型的选择性剪接。 [由RefSeq的,2008年7月提供]
基因本体信息
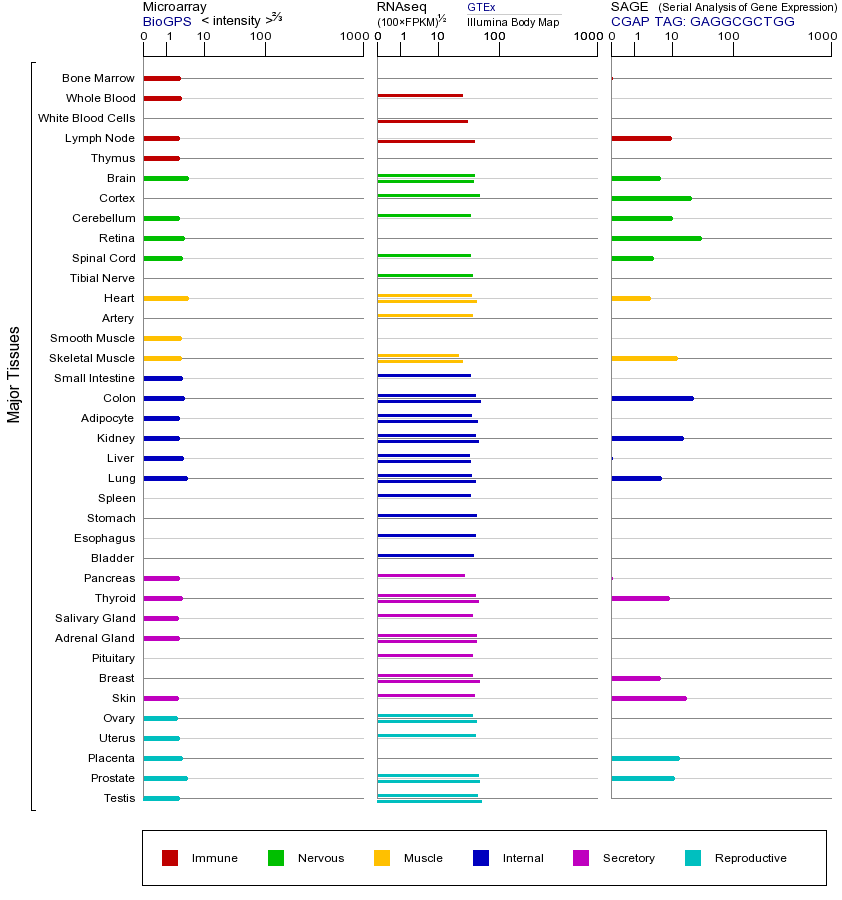
BAD基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
BAD基因的本体(GO)信息:
| 名称 |
|---|
| 4014 Ras signaling pathway [PATH:hsa04014] |
| 4012 ErbB signaling pathway [PATH:hsa04012] |
| 4370 VEGF signaling pathway [PATH:hsa04370] |
| 4024 cAMP signaling pathway [PATH:hsa04024] |
| 4022 cGMP - PKG signaling pathway [PATH:hsa04022] |
| 4151 PI3K-Akt signaling pathway [PATH:hsa04151] |
| 4210 Apoptosis [PATH:hsa04210] |
| 4510 Focal adhesion [PATH:hsa04510] |
| 4910 Insulin signaling pathway [PATH:hsa04910] |
| 4919 Thyroid hormone signaling pathway [PATH:hsa04919] |
| 4722 Neurotrophin signaling pathway [PATH:hsa04722] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5203 Viral carcinogenesis [PATH:hsa05203] |
| 5210 Colorectal cancer [PATH:hsa05210] |
| 5212 Pancreatic cancer [PATH:hsa05212] |
| 5221 Acute myeloid leukemia [PATH:hsa05221] |
| 5220 Chronic myeloid leukemia [PATH:hsa05220] |
| 5218 Melanoma [PATH:hsa05218] |
| 5215 Prostate cancer [PATH:hsa05215] |
| 5213 Endometrial cancer [PATH:hsa05213] |
| 5223 Non-small cell lung cancer [PATH:hsa05223] |
| 5010 Alzheimer's disease [PATH:hsa05010] |
| 5014 Amyotrophic lateral sclerosis (ALS) [PATH:hsa05014] |
| 5152 Tuberculosis [PATH:hsa05152] |
| 5161 Hepatitis B [PATH:hsa05161] |
| 5160 Hepatitis C [PATH:hsa05160] |
| 5145 Toxoplasmosis [PATH:hsa05145] |
| 名称 |
|---|
| Activation of BAD and translocation to mitochondria |
| Activation of BH3-only proteins |
| Adaptive Immune System |
| AKT phosphorylates targets in the cytosol |
| Apoptosis |
| BH3-only proteins associate with and inactivate anti-apoptotic BCL-2 members |
| Cell death signalling via NRAGE, NRIF and NADE |
| Constitutive Signaling by AKT1 E17K in Cancer |
| DAP12 interactions |
| DAP12 signaling |
| Disease |
| Diseases of signal transduction |
| Downstream signal transduction |
| Downstream signaling events of B Cell Receptor (BCR) |
| Downstream signaling of activated FGFR1 |
| Downstream signaling of activated FGFR2 |
| Downstream signaling of activated FGFR3 |
| Downstream signaling of activated FGFR4 |
| Fc epsilon receptor (FCERI) signaling |
| GAB1 signalosome |
| Immune System |
| Innate Immune System |
| Intrinsic Pathway for Apoptosis |
| NGF signalling via TRKA from the plasma membrane |
| NRAGE signals death through JNK |
| p75 NTR receptor-mediated signalling |
| PI-3K cascade:FGFR1 |
| PI-3K cascade:FGFR2 |
| PI-3K cascade:FGFR3 |
| PI-3K cascade:FGFR4 |
| PI3K events in ERBB2 signaling |
| PI3K events in ERBB4 signaling |
| PI3K/AKT activation |
| PI3K/AKT Signaling in Cancer |
| PIP3 activates AKT signaling |
| Programmed Cell Death |
| Role of LAT2/NTAL/LAB on calcium mobilization |
| Signal Transduction |
| Signaling by EGFR |
| Signaling by ERBB2 |
| Signaling by ERBB4 |
| Signaling by FGFR |
| Signaling by FGFR1 |
| Signaling by FGFR2 |
| Signaling by FGFR3 |
| Signaling by FGFR4 |
| Signaling by PDGF |
| Signaling by SCF-KIT |
| Signaling by the B Cell Receptor (BCR) |
| Signalling by NGF |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Prostatic Neoplasms | 0.128715934 | 6 | 0 | BeFree_CTD_human_LHGDN |
| Transient Ischemic Attack | 0.12 | 1 | 0 | CTD_human |
| leukemia | 0.12 | 1 | 0 | CTD_human |
| Platelet mean volume finding | 0.12 | 1 | 1 | GWASCAT |
| Colonic Neoplasms | 0.088173051 | 4 | 0 | LHGDN_RGD |
| Glioma | 0.081357209 | 6 | 0 | BeFree_RGD |
| Hypothyroidism | 0.08 | 1 | 0 | RGD |
| Seizures | 0.08 | 1 | 0 | RGD |
| Gastric ulcer | 0.08 | 1 | 0 | RGD |
| Insulin Resistance | 0.08 | 1 | 0 | RGD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。