AGO2 (argonaute RISC catalytic component 2)
- symbol:
- AGO2
- locus group:
- protein-coding gene
- location:
- 8q24.3
- gene_family:
- Argonaute/PIWI family
- alias symbol:
- hAGO2|Q10|LINC00980
- alias name:
- argonaute 2
- entrez id:
- 27161
- ensembl gene id:
- ENSG00000123908
- ucsc gene id:
- uc003yvm.5
- refseq accession:
- NM_012154
- hgnc_id:
- HGNC:3263
- approved reserved:
- 2000-01-14
AGO2(Argonaute 2)是Argonaute蛋白家族的重要成员,属于RNA诱导沉默复合体(RISC)的核心组成部分。该基因家族在真核生物中高度保守,主要参与RNA干扰(RNAi)和微小RNA(miRNA)介导的基因沉默过程。AGO2通过结合小RNA(如siRNA或miRNA)来识别互补的靶标mRNA,进而引导其降解或翻译抑制,从而调控基因表达。AGO2在多种细胞过程中发挥关键作用,包括发育、细胞分化、免疫应答和病毒防御。其表达或功能异常与多种疾病相关,如癌症、神经退行性疾病和病毒感染。例如,AGO2的突变可能导致RNAi通路缺陷,影响基因调控网络的稳定性,进而促进肿瘤发生或发育异常。在癌症中,AGO2的过表达可能增强致癌miRNA的活性,促进细胞增殖和转移;而表达降低则可能导致抑癌miRNA功能丧失,同样不利于细胞稳态。此外,AGO2还参与抗病毒免疫,某些病毒(如HIV)甚至进化出机制来抑制AGO2功能以逃逸宿主防御。AGO2与其他Argonaute家族成员(如AGO1、AGO3、AGO4)具有相似的结构域(如PAZ和PIWI结构域),但AGO2因其独特的核酸内切酶活性(“Slicer”活性)而尤为突出,能够直接切割靶RNA。该基因家族的共性包括结合小RNA、介导转录后基因沉默以及参与表观遗传调控。研究AGO2的功能和调控机制不仅有助于理解基本的基因沉默原理,还为开发RNAi疗法和疾病治疗策略提供了重要靶点。
This gene encodes a member of the Argonaute family of proteins which play a role in RNA interference. The encoded protein is highly basic, and contains a PAZ domain and a PIWI domain. It may interact with dicer1 and play a role in short-interfering-RNA-mediated gene silencing. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2009]
此基因编码的Argonaute家族从而起到RNA干扰作用的蛋白质的成员。所编码的蛋白质是高度碱性的,并包含一个PAZ域和PIWI域。它可以与DICER1相互作用并在短干扰核糖核酸介导的基因沉默中起作用。已发现该基因编码不同亚型的多个抄本变形。 [由RefSeq的,2009年09月提供]
基因本体信息
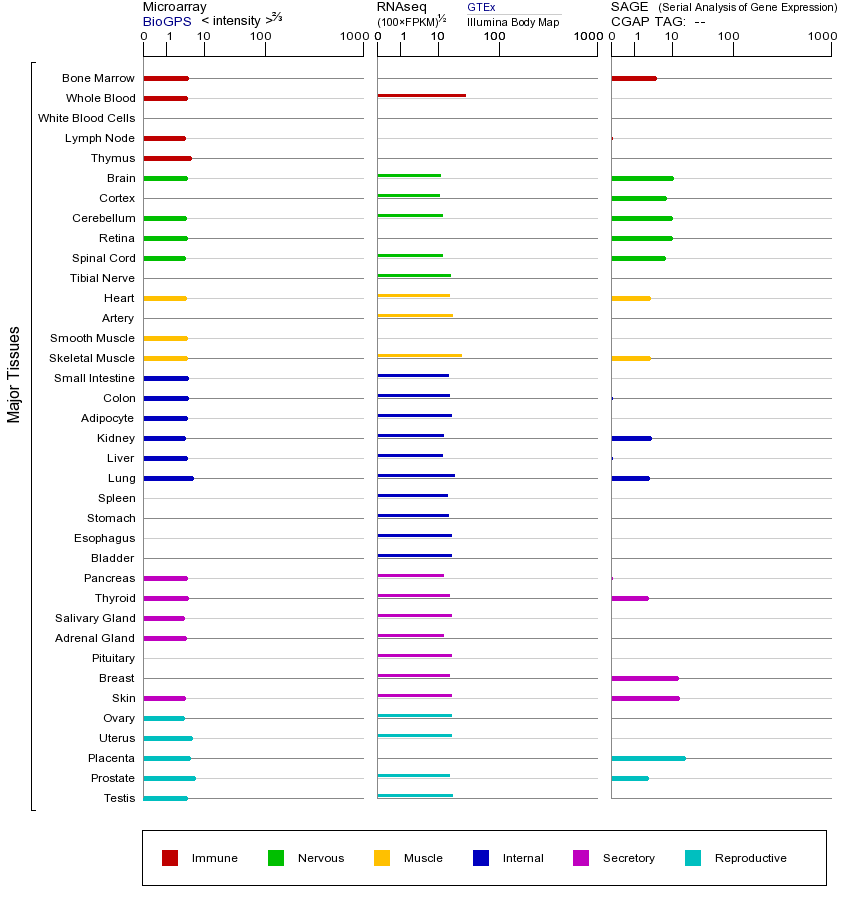
AGO2基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
AGO2基因的本体(GO)信息:
| 名称 |
|---|
| Adaptive Immune System |
| beta-catenin independent WNT signaling |
| Ca2+ pathway |
| DAP12 interactions |
| DAP12 signaling |
| Downstream signal transduction |
| Downstream signaling events of B Cell Receptor (BCR) |
| Downstream signaling of activated FGFR1 |
| Downstream signaling of activated FGFR2 |
| Downstream signaling of activated FGFR3 |
| Downstream signaling of activated FGFR4 |
| Fc epsilon receptor (FCERI) signaling |
| GAB1 signalosome |
| Gene Expression |
| Immune System |
| Innate Immune System |
| MicroRNA (miRNA) biogenesis |
| NGF signalling via TRKA from the plasma membrane |
| PI-3K cascade:FGFR1 |
| PI-3K cascade:FGFR2 |
| PI-3K cascade:FGFR3 |
| PI-3K cascade:FGFR4 |
| PI3K events in ERBB2 signaling |
| PI3K events in ERBB4 signaling |
| PI3K/AKT activation |
| PIP3 activates AKT signaling |
| Post-transcriptional silencing by small RNAs |
| Pre-NOTCH Expression and Processing |
| Pre-NOTCH Transcription and Translation |
| Regulatory RNA pathways |
| Role of LAT2/NTAL/LAB on calcium mobilization |
| Signal Transduction |
| Signaling by EGFR |
| Signaling by ERBB2 |
| Signaling by ERBB4 |
| Signaling by FGFR |
| Signaling by FGFR1 |
| Signaling by FGFR2 |
| Signaling by FGFR3 |
| Signaling by FGFR4 |
| Signaling by NOTCH |
| Signaling by PDGF |
| Signaling by SCF-KIT |
| Signaling by the B Cell Receptor (BCR) |
| Signaling by Wnt |
| Signalling by NGF |
| Small interfering RNA (siRNA) biogenesis |
| Transcriptional regulation by small RNAs |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Myeloid Leukemia | 0.120271442 | 1 | 0 | BeFree_CTD_human |
| Cocaine-Related Disorders | 0.12 | 1 | 0 | CTD_human |
| Mammary Neoplasms | 0.002995792 | 1 | 0 | BeFree_LHGDN |
| Malignant neoplasm of lung | 0.002909916 | 3 | 0 | BeFree_GAD |
| Malignant neoplasm of esophagus | 0.002638474 | 2 | 0 | BeFree_GAD |
| Renal Cell Carcinoma | 0.002367032 | 1 | 0 | GAD |
| Kidney Neoplasm | 0.002367032 | 1 | 0 | GAD |
| Neoplasm Metastasis | 0.001357209 | 5 | 0 | BeFree |
| Liver carcinoma | 0.001357209 | 5 | 0 | BeFree |
| Breast Carcinoma | 0.001085767 | 4 | 3 | BeFree |
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。