编辑推荐:
在CRISPR/Cas9之后,DNA指导的核酸内切酶NgAgo又登上基因组编辑的舞台。几个月来,NgAgo经历了大起大落,一开始备受瞩目,最 近又备受质疑。英国医学研究理事会(MRC)再生医学中心的Pooran Dewari近日调查了NgAgo技术的使用情况。他认为,这种技术还需要更多时间的优化和发展,才能真正与CRISPR正面交锋。
在CRISPR/Cas9之后,DNA指导的核酸内切酶NgAgo又 登上基因组编辑的舞台。几个月来,NgAgo经历了大起大落,一开始备受瞩目,最近又备受质疑。英国医学研究理事会(MRC)再生医学中心的Pooran Dewari近日调查了NgAgo技术的使用情况。他认为,这种技术还需要更多时间的优化和发展,才能真正与CRISPR正面交锋。
NgAgo的独特之处
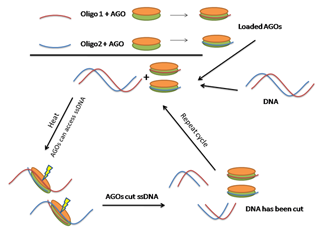
Dewari首先介绍了NgAgo为何引人注目。首先,与Cas9不同,NgAgo不需要PAM序列来识别 目标,这让研究人员有了前所未有的自由,去靶定DNA中的任何序列。其次,NgAgo的特异性是由DNA指导序列决定的,而不是RNA向导。第三,它能够 靶定困难的富含GC区域,这些区域似乎耐受Cas9的切割。
当然,CRISPR/Cas9和NgAgo基因组编辑方法都有自己的优点和缺点。NgAgo DNA向导比较便宜,可自行开展5’-磷酸化或从市场上购买。不过,与RNA向导不同,它不能从质粒中产生。此外,NgAgo/ssDNA的体外组装需要 在55°C孵育,这个温度对哺乳动物细胞很危险。在体内,只有新生的NgAgo蛋白可以与ssDNA向导形成复合物。因此,研究人员必须将5’-P- ssDNA向导与NgAgo表达质粒共转染,才能在体内编辑基因。
NgAgo的真实体验
自今年5月首次发表以来,NgAgo技术一直备受关注,而质粒也被全世界的实验室索要了400多次。研究人 员都在兴奋地试验NgAgo的基因组编辑应用。他们的进展如何呢?Dewari登陆了NgAgo的谷歌论坛,发现许多研究人员都难以形成插入缺失。于是, 他进行了一项调查,询问他们的实验情况,以及与CRISPR/Cas9的比较。
截至2016年8月1日,总共165名研究人员答完了调查问卷。当被问及他们是否能用NgAgo形成插入缺 失时,在88名受访者中只有1人证实,在目标位点成功形成了插入缺失。同时,41名受访者试图利用此技术进行表位标记,但只有1人能够在目标位点检测到标 记的插入(knock-in)。总的来说,64%的研究人员对新方法不满意,65%的人希望有人能优化NgAgo的操作。绝大多数的受访者(70%)是利 用Lipofectamine方法将NgAgo导入细胞的,与最初的文章相同。
Dewari认为,尽管大多数受访者都在努力挣扎着,但这并不意味着NgAgo没有作用。从好的方面来看,少数受访者的确实现了成功的插入缺失和表位插入,这表明NgAgo能在哺乳动物细胞中实现基因组编辑。
然而,大多数研究人员的现状表明,NgAgo系统可能不容易上手。原因之一是所谓的5’-磷酸化ssDNA 向导的不稳定性,它可能被细胞机制迅速降解,使得细胞中只有少量的NgAgo/ssDNA复合物。正如原文所提到的,为了实现更好的效果,研究人员需要将 ssDNA反复转染。另一个可能的原因是成熟的NgAgo蛋白无法结合ssDNA向导。在翻译过程中,也许只有小部分的新生NgAgo蛋白能够与 ssDNA形成有功能的复合物,导致下游的编辑效率不高。
总的来说,Dewari认为我们还需要更多的时间来优化NgAgo,才能得出任何主要结论。他建议从事相关 工作的研究人员在谷歌论坛、Twitter或Addgene的博客上讨论,将研究成果与大家分享。基因组编辑领域一直在飞速发展,说不定哪天就有人开发出 NgAgo的突变蛋白或全新的核酸内切酶。
附上韩老师 NgAgo技术的详细操作步骤:
A general protocol of NgAgo/gDNA-mediated genome editing
1. Cell culture
293T cells are maintained in high-glucose DMEM (Gibco) supplemented with 10% FBS (Gibco) and penicillin/streptomycin at 37°C with 5% CO2 incubation. When cells reach their ≈60% confluence, perform transfection. Before transfection, cells are changed to medium containing 2% FBS(heat inactivated). Since 293T cells are not firmly attached, be gentle and do not disturb cells when changing medium.
2. Transfection
2-1 NLS-NgAgo expressing plasmid is extracted with Wizard® Plus SV Minipreps DNA Purification System (Promega), and is adjusted to 100 ng/μl in water (pH 8.0, alkalization by NaOH).
2-2 5’ phosphorylated ssDNA guides are dissolved to 100 ng/μl in water (PH 8.0) For each well of a 24-well plate, 200-250 ng NLS-NgAgo expression plasmid and 100-300 ng guides are diluted in 50 μl Opti-MEM (Gibco); 1.25 μl Lipofectamine® 2000 is diluted in 50 μl Opti-MEM. Incubate the DNA mix and lipofectamine mix for 5 min.
2-3 Combine the DNA mix and lipofectamine mix with gentle pipetting and incubate for 20 min. The DNA/lipo mixture is then added into each well of cells.
*Since NgAgo follows “one-guide faithful” rule, i.e. guides can only be loaded when NgAgo protein is in the process of expression, to improve the efficiency of gDNA loading to NgAgo, sometimes multiple transfection of gDNA helps (e.g. depending on the different kinetics of NgAgo expression in different cells, a second transfection of gDNA can be performed 8, 12 or 24 hours after the primary transfection)
*As stated below, cells will be harvested 48-60 hours after transfection. 90% confluence of the cells on harvesting is ideal. Cell overplating significantly weakens the efficacy of genome editing. Taking HDR as an example, it occurs only during S and G2 phases.
3. Genomic DNA extraction
3-1 48-60 hours after transfection, cells are harvested by trypsin digestion. Four wells of cells are combined into a 1.5 ml EP tube.
3-2 For genomic DNA extraction, 500 μl of cell lysate buffer (50 mM Tris,100 mM EDTA,0.5% SDS,pH 8) and 10 μl proteinase K (10 mg/ml)are added into each tube and mixed gently and sufficiently. Bathe the tubes at 55℃ for 2 hours.
3-3 200 μl Tris-Phenol and 200 ul trichloromethane are added into each tube and mixed gently and sufficiently. After incubation for 5 min, samples are spun at 12,000 rpm for 15 min to separate aqueous phase from Phenol phase.
3-4 Carefully collect the aqueous phase into a clean EP tube.
3-5 Repeat the Steps 3-3 and 3-4 once and pool the aqueous phase.
3-6 Add 500 μl trichloromethane into the collected aqueous phase, mix gently and sufficiently, stay for 5 min, and then spin the sample at 12,000 rpm for 15 min to separate aqueous phase from Phenol phase. Carefully remove the aqueous phase into a clean EP tube.
3-7 Repeat the Steps 3-6 once.
3-8 Add 900 μl EtOH to the collected aqueous phase and incubate at -20°C for 30 min.
3-9 Centrifuge the sample at 12,000 rpm for 10 min, and then the DNA sediment is washed with 500 μl 75% EtOH thrice.
3-10 Air-dry the DNA sediment, add 50 μl 0.5 x TE and then adjust the genome DNA to 100 ng/μl for later use.
Note:
1. NgAgo/gDNA system is extremely sensitive to contamination of intracellular bacteria (including mycoplasma) which are widespread and leave no visible signs of presence. Carefully excluding the presence of intracellular bacteria before performing experiments.
2. Because Agos need magnesium, avoid EDTA when detaching and seeding cells into the plates for transfection. Alternatively, supplementing Mg2+ to 5 mM (may need optimization to your cell type).
3. Avoid using Lipofectamine® 3000 since the supplement P3000 interferes with ssDNA transfection. Lipofectamine® 2000 is a good choice. Other transfection methods such as electroporation are yet to be tested.
4. Ideally, 5' phosporylation of ssDNA guide by using T4 PNK (Biolab):
T4 PNK 2 μl
T4 ligase buffer (containing ATP): 12 μl
1OD ssDNA (about 33 μg) in H2O: 100 μl
(alternatively) additional ATP (25 mM) 2 μl
Add H2O to a final volume of 120 μl
Incubate at 37℃ overnight
After 5' phosphorylation, the resultant ssDNA needs no purification, dilute by water (PH8) to 300 μl with a final concentration of 10 nM or 100 ng/μl)
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。


